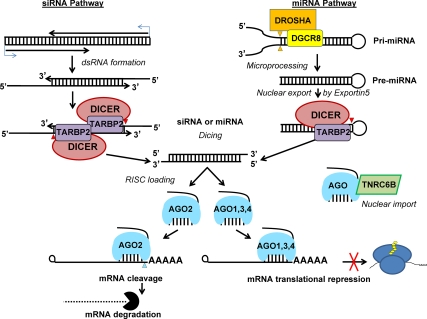
Fig. 4.
siRNA and miRNA biosynthesis. siRNA are initially transcribed from promoters within viral genomes, retrotransposons, adjacent genes with a tail-to-tail or overlapping head-to-head (data not shown) orientation of RNA promotion, or from pseudogene mRNA with antisense promotion paired to their functional gene paralogs. The resulting transcripts form a region of dsRNA. By contrast miRNA are transcribed from RNA pol II promoters intronic to other protein-encoding genes or under their own promoters and fold into a hairpin structure containing a region forming a dsRNA stem called a primary miRNA (pri-miRNA). The Microprocessor complex, composed of nuclear type III RNA endonuclease RNASEN/DROSHA and its cofactor DGCR8, excises the pre-miRNA by cleaving at the base of the pri-miRNA stem (orange triangles) and exported from the nucleus by exportin 5. Subsequently, the cytoplasmic type III RNA endonuclease DICER and cofactor TAR RNA binding protein 2 (TARBP2) cleaves the 5′-overhangs of the siRNA precursor or the loop of the pre-miRNA (red triangles). A mature siRNA or miRNA (single strand of the dsRNA) is loaded onto the Argonaute-containing effector complex, RNA-induced silencing complex. In Argonaute cocrystal structures the seed sequence is held rigidly between the PIWI and PAZ domains of the Argonaute allowing base pairing required for targeting. In mammals, AGO2 contains the conserved residues necessary for and demonstrated activity of endonucleolytic cleavage of target mRNA. After mRNA endonucleolytic cleavage, the target mRNA is decapped and exonuclease digestion occurs. The remaining Argonautes (AGO1, AGO3, and AGO4) lack the conserved endonuclease residues but may participate in the alternative outcome of miRNA action, translational repression.