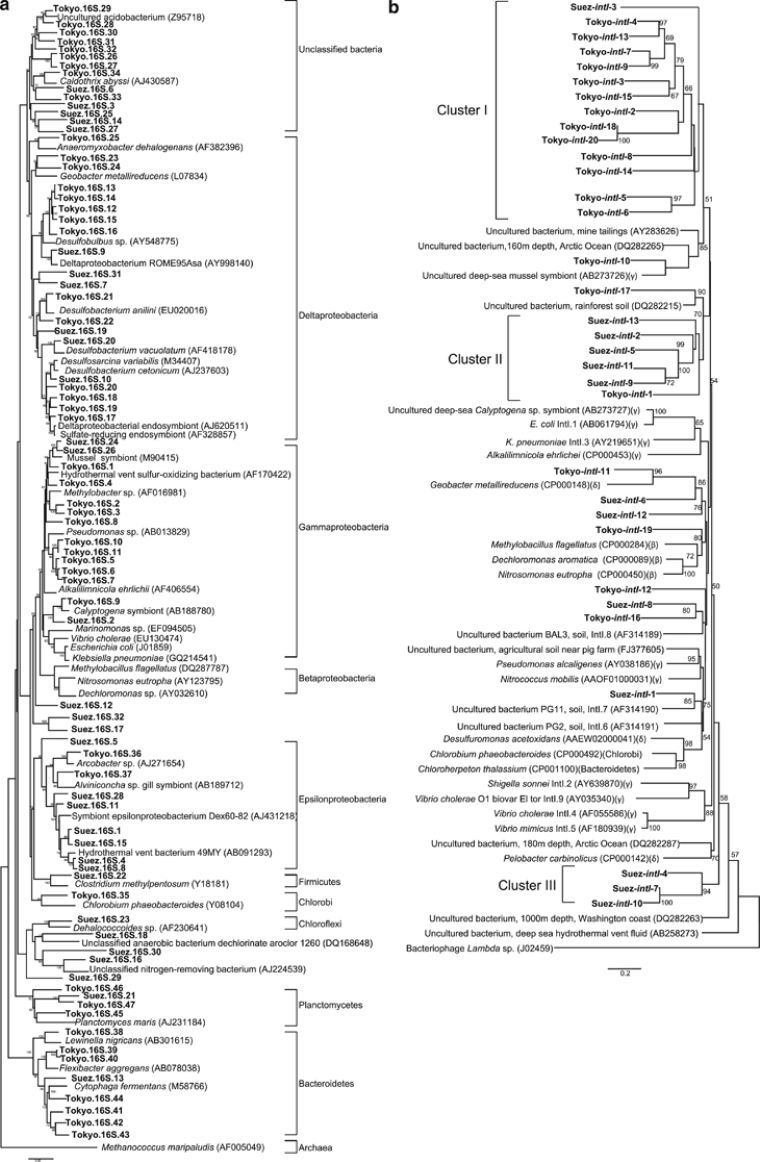
Figure 2.
Consensus trees were constructed based on 16S rRNA gene nucleotide sequences (a) and intI deduced amino-acid sequences (b). The trees showed the phylogenetic relationship between the recovered sequence phylotypes and their homologues from database. The trees were constructed by neighbor-joining analyses and confirmed by maximum-parsimony and maximum-likelihood methods. Bootstrap values of >50% were indicated at the branch roots. Recovered sequences were highlighted in bold. Abbreviations (β), (γ) and (δ) represented the classes Betaproteobacteria, Gammaproteobacteria and Deltaproteobacteria, respectively. The bars represented 0.05 and 0.2 changes per a nucleotide and an amino acid, respectively.