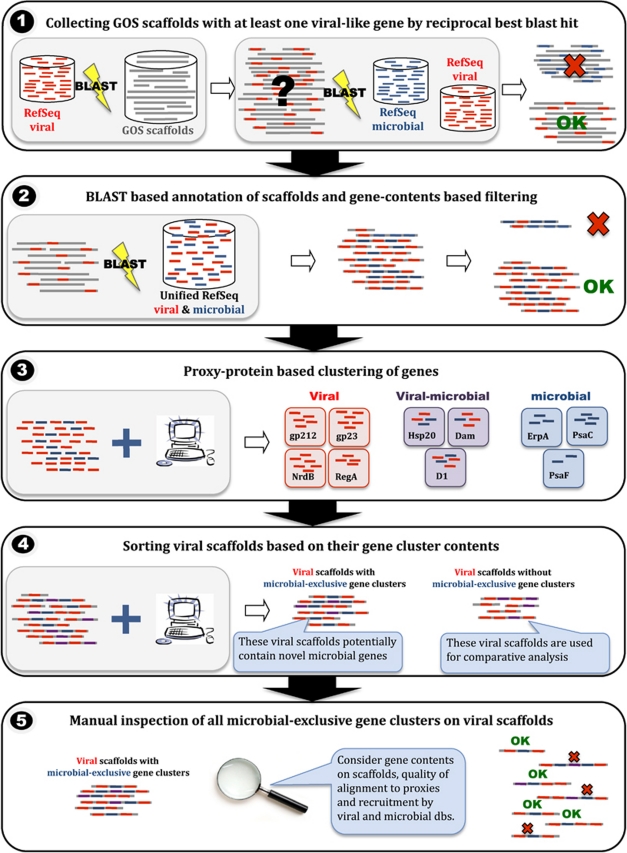
Figure 1.
Pipeline for the identification of viral scaffolds with microbial genes. (1) Candidate scaffolds containing at least one potential viral gene were identified using a bi-directional BLAST (Basic Local Alignment Search Tool) search of all proteins in the RefSeq-viral database of the National Center for Biotechnology Information against all GOS scaffolds, and then a BLAST search of all significant best hits against a combined RefSeq-viral and RefSeq-microbial proteins database. Scaffolds with best hits from RefSeq-viral were further considered. (2) Gene contents of all candidate scaffolds were determined using an iterative sequence similarity-based procedure against the combined dataset of RefSeq-viral and RefSeq-microbial proteins. (3) Genes were clustered based on sequence similarity of their RefSeq hits, with each cluster being tagged based on the origin of its RefSeq proteins (viral exclusive, microbial exclusive or viral microbial). For the purpose of clustering we consider all genes' proxy proteins rather the genes and their scaffolds. Scaffold tagging was determined based on gene contents and on recruitment against the Northern Line Islands datasets (Figure 2). (4) Viral scaffolds were sorted into scaffolds containing members from viral or viral-microbial clusters only, and scaffolds also containing members of microbial exclusive clusters. Purple genes refer to genes that belong to viral-microbial clusters, red and blue refer to members of viral-exclusive and microbial-exclusive clusters, respectively. (5) The latter set was inspected manually in order to validate their origin and annotation.