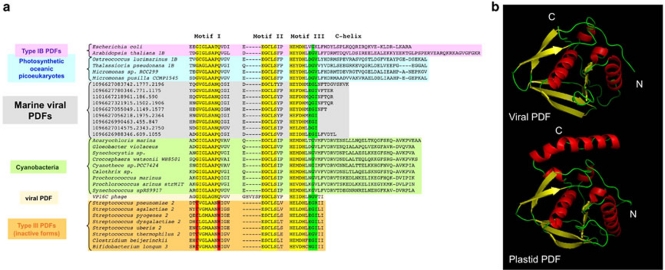
Figure 3.
Marine viral PDFs do not display the conserved C-terminal helix of subtype IB PDF and constitute a new class of active PDFs. (a) A phylogenetic tree was constructed from an alignment of ∼200 PDFs recapitulating sequence and phylogenetic diversity (Supplementary Figure S2). Among marine viral PDFs, only representative members are displayed (colored in gray). These proteins are related to PDFs from cyanobacteria (shown in green) and photosynthetic planktonic picoeukaryotes (shown in blue). Proteins showing the closest similarities around the three conserved motifs 1, 2 and 3 (colored in yellow) and C-helix were selected and realigned, and the motifs are shown. Unlike type III inactive PDFs (colored in orange below), all required residues of the motifs are conserved, which is strongly suggestive of peptide deformylase activity. The closest structural models (that is, E. coli and Arabidopsis thaliana PDF1B) are indicated on top. In both cases, the C-terminus folds as a α helix. (b) A refined three-dimensional model for viral PDF (top) compared to thethree-dimensional crystal structure of the most relevant PDF (see panel a) from chloroplastic PDF1B (PDB code 3cpmA; bottom). The two structures are shown in the same orientation, that is, toward the entry of the active site crevice. Both N- and C-ends are indicated in white with N and C, respectively.