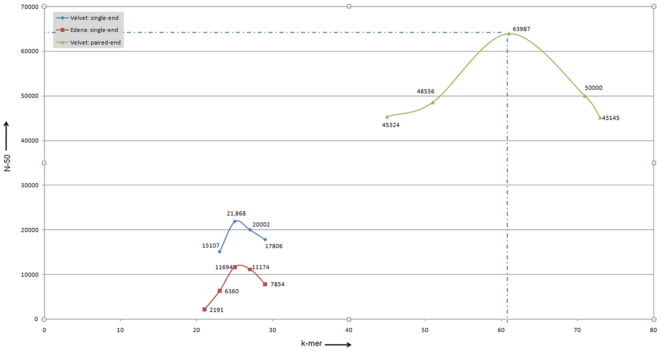
Figure 1. De novo assembly of C. concisus UNSWCD genome.
k-mer values ranging from 19–33 were used for assembling the single-end read data for UNSWCD strain. The Velvet assembler generated a maximum N-50 value of 21,868 with a genome coverage of 1.68 Mb (352 contigs) where k = 25. The Edena assembler produced a bigger (1.79 Mb) but more fragmented assembly (459 contigs) with an overlap length of 25. Genome assembly was generated using a set of paired-end reads and Velvet assembler; this resulted in an improved base coverage (1.8 Mb) as well as a much improved N-50 value of 63,987 for a k-mer value of 61.