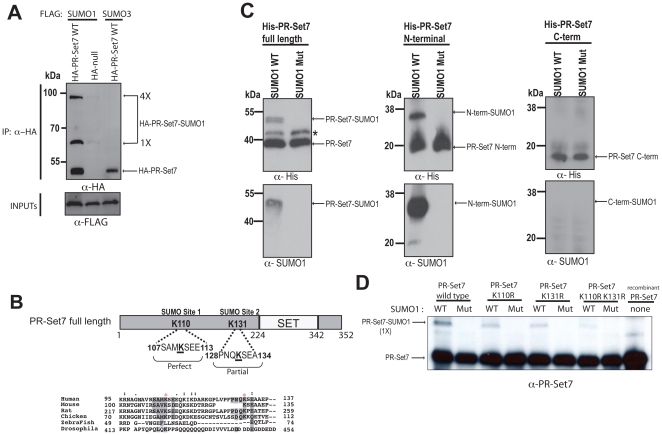
Figure 4. PR-Set7 is selectively modified by SUMO1 in cells and SUMOylated at K110 or K131 in vitro.
(A) HEK 293 cells were co-transfected with the indicated expression plasmids. Cell extracts were prepared in the presence of NEM 2 days post-transfection, followed by immunoprecipitations using an anti-HA antibody. HA-bound samples were fractionated by SDS-PAGE prior to Western analysis using anti-FLAG or anti-HA antibodies. (B) Two putative PR-Set7 SUMOylation sites, one perfect and one partial, were identified using SUMOsp 2.0 [40]. ClustalX was used to align the sequence surrounding the putative SUMO sites of PR-Set7 in various organisms. Conserved residues are shaded and red asterisks mark K110 and K131. (C) Recombinant wild type PR-Set7, N-terminal (aa1–191) or C-terminal (aa192–352) fragments were used as substrates in in vitro SUMOylation assays. Reactions were fractionated by SDS-PAGE gel followed by Western analysis using anti-His and anti-SUMO1 antibodies. Asterisk (*) denotes an unknown contaminating protein. (D) Recombinant wild type PR-Set7, K110R, K131R or K110/131R double mutant were used as substrates in in vitro SUMOylation assays. Reactions were fractionated by SDS-PAGE followed by Western analysis using anti-PR-Set7 antibodies.