Figure 6.
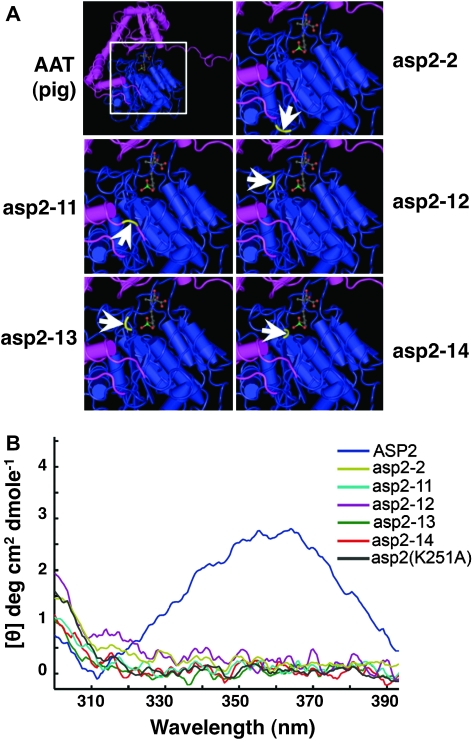
Amino Acid Substitutions in asp2 Suppressors Are Located in the PLP Binding Pocket.
(A) The site of asp2 suppressor mutation amino acid changes mapped onto the crystal structure model of the swine cytosolic AAT enzyme. The PLP cofactor is visible as a ball-and-stick structure in the center of the protein. Each mutant protein version is represented on a close-up of the swine AAT enzyme, as illustrated by the white box. Arrows indicate the positions for the mutant residues highlighted in yellow.
(B) Only the WT ASP2 has a CD-spectrum indicative of a having covalently bound PLP cofactor. Near UV circular dichroic (CD) spectrum data for WT ASP2 and various mutant proteins are shown.