SUMMARY
The obligate intracellular bacterial pathogen Chlamydia trachomatis injects numerous effector proteins into the epithelial cell cytoplasm to manipulate host functions important for bacterial survival. In addition, the bacterium secretes a serine protease, chlamydial protease-like activity factor (CPAF). Although several CPAF targets are reported, the significance of CPAF-mediated proteolysis is unclear due to the lack of specific CPAF inhibitors and the diversity of host targets. We report that CPAF also targets chlamydial effectors secreted early during the establishment of the pathogen-containing vacuole (“inclusion”). We designed a cell-permeable CPAF-specific inhibitory peptide and used it to determine that CPAF prevents superinfection by degrading early Chlamydia effectors translocated during entry into a pre-infected cell. Prolonged CPAF inhibition leads to loss of inclusion integrity and caspase-1-dependent death of infected epithelial cells. Thus, CPAF functions in niche protection, inclusion integrity and pathogen survival, making the development of CPAF-specific protease inhibitors an attractive anti-chlamydial therapeutic strategy.
INTRODUCTION
The obligate intracellular bacterial pathogen Chlamydia trachomatis primarily infects epithelial cells of the urogenital tract and the conjunctiva (Adderley-Kelly and Stephens, 2005) that can lead to severe complications such as pelvic inflammatory disease, ectopic pregnancy and infertility. Similarly, ocular infections can lead to trachoma, the leading cause of infectious blindness worldwide (Hu, 2010). Infection is initiated by attachment and entry of elementary bodies (EB), the infectious form of Chlamydia, into epithelial cells. Upon entry, EBs transition into replicative reticulate bodies (RB) and establish a parasitophorous vacuole (“inclusion”) that prevents fusion with lysosomal compartments. At mid- to late stages of infection, RBs revert back to EBs, and are released to infect neighboring cells (Belland et al., 2003).
C. trachomatis employs a type III secretion (T3S) system to translocate virulence proteins directly into the cytoplasm of the host cell. These effector proteins mediate cell invasion, rerouting of membrane transport, and manipulation of signaling pathways that are important in immunity (Valdivia, 2008). More than 5% of Chlamydia’s genome is predicted to encode T3S effector proteins (Arnold et al., 2009), including a family of inclusion membrane proteins (Incs) that reside at the interface of the host cytoplasm and inclusion membrane (Scidmore-Carlson et al., 1999). These proteins are implicated in the modulation of various host cellular functions, including membrane transport, apoptosis and microtubule dynamics (Betts et al., 2009). Unfortunately, because of Chlamydia’s obligate intracellular lifestyle and lack of tools for genetic manipulation, most putative chlamydial effectors remain poorly characterized.
In addition to T3S effectors, proteins that use the general secretion pathway can access the host cell cytoplasm to manipulate host cellular processes (Chen et al., 2010). A prominent example is the chlamydial protease-like activity factor (CPAF), a serine protease that is first secreted into the inclusion lumen and eventually crosses the inclusion membrane (Zhong et al., 2001). CPAF cleaves transcription factors (RFX5 and USF1) required for the expression of antigen presentation molecules (MHC) (Zhong et al., 2001), the pro-apoptotic factors Bim and Puma (Pirbhai et al., 2006; Zhong et al., 2001) and p65/RelA, a transcription factor required for NF-κB signaling (Christian et al., 2010). Recently, there has been an expanding list of identified CPAF targets, underscoring the multiple roles played by this protease in the biology of Chlamydia infections. CPAF cleaves the adherence junction protein nectin1 (Sun et al., 2008), the MHC-like protein CD1d (Kawana et al., 2007), the pro-inflammatory protein HMGB1 (Yu et al., 2010), the mitotic cell cycle regulator Cyclin B, and PARP – a mediator of DNA-damage during apoptosis (Paschen et al., 2008). CPAF also remodels intermediate filaments (Dong et al., 2004; Kumar and Valdivia, 2008) that circumscribe the inclusion to maintain its integrity and limit the exposure of inclusion contents to cytoplasmic microbial pattern recognition receptors (Kumar and Valdivia, 2008).
Recent observations with an inducible CPAF over-expression system suggested that this protease can initiate a host cell death pathway that mimics the necrotic cell death observed at the end of the chlamydial life cycle (Paschen et al., 2008) and thus contribute to acute inflammation and tissue scarring. Yet the significance of CPAF-mediated proteolysis during infection is unclear due to the lack of specific CPAF inhibitors and the large number and variability in host cell targets. Nonetheless, the abundance of specific targets implies that CPAF-mediated proteolysis represents a core strategy employed by Chlamydia to modify host-signaling pathways and usurp the cellular machinery for its own benefit.
Here we report that in addition to targeting host proteins, CPAF cleaves bacterial effectors translocated during invasion and early in inclusion biogenesis. We demonstrate that CPAF is essential for bacterial replication, maintaining inclusion structural integrity and evasion of caspase-1-dependent cell death in epithelial cells. These results establish CPAF as a virulence factor required for Chlamydia survival within infected cells and reveal unexpected roles for this protease in regulating T3S effectors after their translocation into the host cell and in maintaining host cell viability.
RESULTS
A subset of Chlamydia effector proteins is sensitive to proteolysis
Approximately 10% of the Chlamydia genome encodes proteins that potentially access the cytoplasm of the infected host cell and act as effectors (Arnold et al., 2009) (Saka and Valdivia, unpublished). Because effectors are secreted at distinct stages during infection, we hypothesized that their activity in the host cytoplasm is regulated as inappropriate activity of effectors might negatively impact bacterial growth. Since proteolysis is a common mechanism of post-translational control, we tested a panel of 309 recombinant Chlamydia-specific proteins, including known and potential effector proteins, for sensitivity to proteolysis following incubation with lysates from infected and uninfected HeLa cells (Figure S1). Thirty of the expressed chlamydial proteins were sensitive to cytosols derived from infected cells (Figure S1A). Protein processing ranged from the generation of distinct cleavage fragments to complete degradation (Figure S1B). The protease-sensitive chlamydial proteins comprised inclusion membrane proteins (n=11), outer membrane proteins (n=3), proteases (n=2) and ORFs of unknown function (n=14) (Figure S1C and Table S2). Thirteen of the thirty were also sensitive to proteolysis when treated with lysates from uninfected cells, indicating that these proteins are likely targets of host proteases (Figure 1A, Figure S1C–D and Table S2).
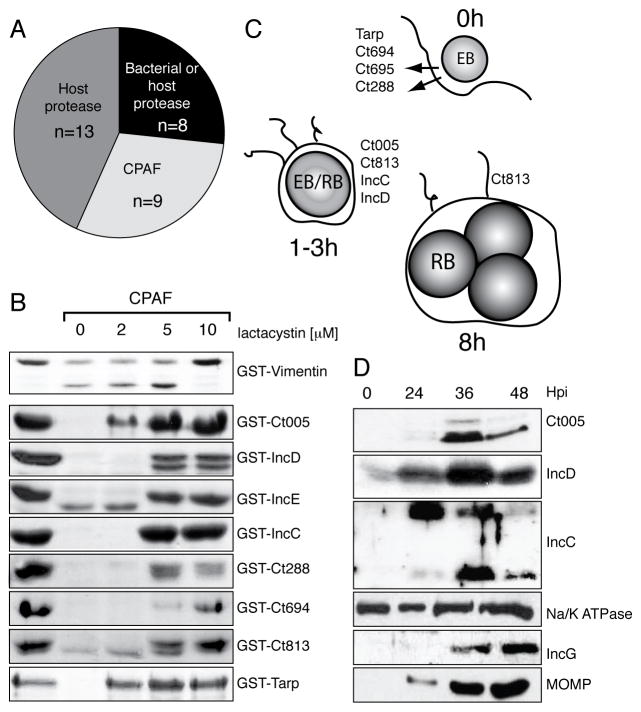
Figure 1. CPAF cleaves a subset of early chlamydial effectors.
(A) Recombinant Chlamydia ORFs (n=309) were tested for sensitivity to host – and bacterial – derived proteases.
(B) Recombinant CPAF cleaved chlamydial substrates in a cell-free proteolysis assay. Approximately 40 ng purified his-tagged CPAF was incubated with 48μg GST-tagged Chlamydia proteins at 37°C for 20 min with increasing concentrations of lactacystin. Cleavage products were run on an SDS PAGE gel and visualized with Coomassie.
(C) Schematic summary of bacterial targets of CPAF. CPAF specifically cleaves chlamydial effectors translocated during EB entry and early inclusion biogenesis, with the exception of Ct813, which is transcribed starting at 8hpi.
(D) The steady state levels of the Inc CPAF substrates decrease at late stages of infection. Membranes from Chlamydia-infected HeLa cells were harvested at 0, 24, 36 and 48 hpi, purified, and the abundance of the chlamydial proteins Ct005, IncC and IncD monitored with specific antibodies. IncG and MOMP are an inclusion membrane protein and an outer membrane protein, respectively, that were determined not to be protease sensitive. Na/K ATPase serves as host membrane loading control.
The secreted protease CPAF cleaves a subset of effector proteins
Cleavage of nine chlamydial proteins by was inhibited by pre-treatment of infected HeLa cytosols with lactacystin but not the unrelated proteosomal inhibitors MG132 and ALLN (Figure S1D and data not shown). Because this inhibitor sensitivity profile is shared by CPAF (Pirbhai et al., 2006; Zhong et al., 2000) we tested if CPAF played a role in these cleavage events by treating cytosols from infected HeLa cells with anti-CPAF antisera. Anti-CPAF antisera, but not control antisera, blocked the degradation of recombinant bacterial proteins (Figure S2A). In this manner we determined that all proteolysis-sensitive Chlamydia proteins were likely substrates of CPAF (Figure 1A). We then tested if CPAF was sufficient for these cleavage events. Recombinant CPAF readily cleaved vimentin, a well-characterized CPAF substrate (Kumar and Valdivia, 2008; Paschen et al., 2008) and GST-tagged chlamydial proteins Ct005, IncD (Ct115), IncE (Ct116), IncC (Ct233), Ct288, CT694, CT695, Ct813 and TARP (Ct456) (Figure 1B). In contrast, GST-tagged proteins that were identified as (1) not sensitive to proteolysis, (2) sensitive to a non-CPAF chlamydial protease or (3) sensitive to a host protease were not cleaved by recombinant CPAF (Figure S1D, S2B, and data not shown). Like endogenous CPAF, proteolysis by recombinant CPAF was inhibited by lactacystin but not by MG132, ALLN, or a range of serine protease inhibitors (Figure 1B and FigureS2C). The proteins identified as potential CPAF substrates all contain putative T3S signals (Arnold et al., 2009), and fall into two main categories: (1) experimentally validated effectors that are pre-packed into EBs (Clifton et al., 2004; Hower et al., 2009) and (2) inclusion membrane proteins whose mRNAs are expressed early (1–3 h) after invasion (Belland et al., 2003) (Figure 1C).
Figure 2. A cell-permeable CPAF-specific inhibitory peptide blocks cleavage of Chlamydia effectors during infection.
(A–C) Tarp translocated by EBs is a target of CPAF-mediated degradation. Uninfected HeLa cells and HeLa cells pre-infected with L2 for 30 h were treated with a cell permeable poly-arginine tagged CPAF inhibitory (“anti-CPAF”) peptide and infected with EBs for 10 min. Tarp translocation was indirectly visualized with an anti-phosphotyrosine antibody (A) and quantified in 50 separate cells (B). Error bars represent +/− St. error. The stability of newly translocated Tarp was assessed by infecting cells as in (A) but with S35-labeled EBs, followed by sequential immunoprecipitation of Tarp and MOMP and PhosphoImager analysis of precipitated material (C). Note CPAF-dependent Tarp degradation in pre-infected HeLa cells. Error bars represent +/− SD.
(D) Inhibition of CPAF activity by inhibitory peptide. IC50 values were assessed by monitoring the cleavage of a synthetic fluorophore-tagged, vimentin-derived substrate by HPLC. Assays were performed in the presence of increasing amount of inhibitors. The IC50 values of the anti-CPAF peptide and Lactacystin were 0.05μM +/− 0.007 sd and 10μM +/− 2.3 sd, respectively.
(E–F) Anti CPAF peptide, but not a scrambled control peptide, broadly inhibits degradation of CPAF substrates in vitro and in vivo. CPAF cleavage of GST-chlamydial substrates were performed as in Fig. 1B but in the presence of anti-CPAF or scrambled control peptide (E). For in vivo inhibition effects on Chlamydia (L2), infected HeLa cells were treated with peptide at 12 hpi, and harvested at 30 hpi (F). Vimentin and Puma cleavage is inhibited in infected cells treated with anti-CPAF peptide.
(G–H) CPAF restricts EB entry into pre-infected cells. Cells were infected as in (A), except that the secondary infections (30 min) were performed with fluorescently-labeled EBs (green) and infected cells were not permeabilized. Extracellular EBs were distinguished from intracellular EBs (green) based on their immunoreactivity to anti-L2 antibodies (red). Extracellular bacteria appear yellow (G). Also note clustering of internalized bacteria at a perinuclear site. Number of internalized EBs was quantified per cell (H). Representative of two experiments performed in duplicate (n=40 cells). Error bars represent +/− St. error.
We next addressed whether the chlamydial CPAF substrates were cleaved during infection. First, we expressed the EGFP-tagged CPAF substrates Ct005 and IncD in HeLa cells and determined that these proteins were processed only upon Chlamydia infection (Figure S2D). We generated antibodies against three of these proteins (Ct005, IncC and IncD) and confirmed by immunofluorescence microscopy (IF) that they are expressed by 6 h post-infection (Figure S2E). Because CPAF is not expressed until later in infection (>18 h), we predicted that if Ct005, IncC and IncD were CPAF substrates their abundance would decrease with the onset of CPAF secretion. We assessed protein abundance semi-quantitatively by immunoblot analysis of membranes isolated from Chlamydia-infected HeLa cells. The levels of major outer membrane protein (MOMP) and the inclusion membrane protein IncG, which are not CPAF substrates, increased throughout infection, reflecting the increased bacterial loads in these cells. In contrast, the levels of Ct005, IncC and IncD increased from 24–36 h but dropped at 48 h (Figure 1D). Overall, these experiments suggest an inverse correlation between the onset of CPAF expression and the abundance of these three secreted chlamydial proteins. Consistent with their potential as bona fide CPAF targets, endogenous Ct005, IncC and IncD from isolated membranes of Chlamydia-infected cells (24 hpi) were efficiently cleaved by recombinant CPAF in vitro (Figure S2F).
CPAF cleaves Tarp during Chlamydia entry into pre-infected cells
We identified the EB proteins Tarp and Ct694 as potential substrates of CPAF-mediated degradation (Figure 1B–C). Tarp and CT694 are pre-packaged into EBs and translocated into the host cell during chlamydial invasion (Clifton et al., 2004; Hower et al., 2009). Recombinant CPAF specifically degraded Tarp and Ct694 from EB lysates, but not housekeeping proteins, and its incubation with lysates did not result in the non-specific degradation of EB proteins (Figure S2G–H).
One scenario where Tarp and Ct694 would encounter CPAF would be when an EB infects a cell that already contains a mature inclusion. We predicted that under these conditions Tarp translocated by EBs would be rapidly degraded. To test this, we first compared the levels of Tarp at EB attachment sites on uninfected versus pre-infected HeLa cells. Tarp is phosphorylated at multiple tyrosine residues by host tyrosine kinases (Jewett et al., 2008; Mehlitz et al., 2008) and IF staining with anti-phosphotyrosine antibodies reveals a prominent cup of immunoreactive material at EB attachments sites (Clifton et al., 2004). We observed multiple phosphotyrosine-positive foci immediately adjacent to EBs intimately attached to the plasma membrane of HeLa cells. These foci, however, are largely absent at EB attachment sites in HeLa cells that were pre-infected for 30 h (Figure 2A–B), indicating that Tarp translocation or phosphorylation is inhibited or that translocated Tarp is degraded. To determine the stability of translocated Tarp under these infection conditions, we infected HeLa cells or inclusion-containing HeLa cells (30 hpi) with 35S-radiolabelled EBs and immunoprecipitated Tarp at 10 min post infection. Radiolabeled Tarp, but not the outer membrane protein MOMP, was degraded in HeLa cells harboring mature inclusions but not in uninfected HeLa cells (Figure 2C). Overall, these experiments indicate that EB-associated effectors, like the early inclusion membrane proteins described above, are subject to degradation in a manner that correlates with the presence of CPAF.
A cell-permeable CPAF-specific inhibitory peptide prevents CPAF cleavage of host and Chlamydia substrates
Because the lack of a system for genetic manipulation of Chlamydiae precludes the generation of CPAF-deficient mutants or the mutation of CPAF cleavage sites in effectors, it is difficult to directly test the functional consequence of these proteolytic events. However, recent structural information on CPAF (Huang et al., 2008) provided the basis for the design of a CPAF-specific inhibitory peptide (SLFYSPMVPHFWAELRNHYATSGLK) that occludes the active site of the mature enzyme. This CPAF peptide258–283 is removed during auto-catalytic activation of the zymogen. As a control, we synthesized a scrambled version of the peptide. To facilitate delivery and uptake into mammalian cells we included an N-terminal nine-Arg tail on both peptides (Stewart et al., 2008). We tested the efficiency of the poly-Arg-tagged CPAF inhibitory peptide in vitro using an HPLC-based enzyme assay that measures the cleavage of an Abz-tagged Vimentin derived peptide (Abz-VRLRSSVPGV). The “anti-CPAF peptide” blocked CPAF-mediated proteolysis with ~100 fold higher efficiency than lactacystin, the only known inhibitor of CPAF (Figure 2D). The anti-CPAF peptide, but not the scrambled control peptide, also inhibited cleavage of GST-tagged chlamydial substrates and GST-Vimentin in vitro (Figure 2E and data not shown). Importantly, the anti-CPAF peptide inhibited cleavage of Vimentin and Puma within infected epithelial cells (Figure 2F), indicating that mammalian cells efficiently internalize the peptide and that CPAF activity can be inhibited within live, infected cells. Moreover, the CPAF inhibitory peptide did not induce any obvious toxicity or alter proteasome function in uninfected cells (data not shown).
Next, we tested if this inhibitory peptide could reverse Tarp degradation upon secondary infections. Indeed, anti-CPAF peptide restored the accumulation of Tyr-phosphorylated proteins at EB attachment sites on pre-infected cells (Figure 2B) and partially blocked the degradation of Tarp in pulse-chase experiments (Figure 2C), suggesting that Tarp may be a target for other host or bacterial proteases. Overall, these observations strongly suggest that early Chlamydia effectors are in vivo targets of CPAF and that a CPAF-specific inhibitory peptide can efficiently inhibit CPAF activity both in vitro and in vivo.
We postulated that CPAF-mediated degradation of effectors secreted by EBs during invasion may protect pre-infected cell against superinfection. To test whether infected cells are refractory to re-infection, we quantified the number of EBs internalized by uninfected cells and pre-infected cells. We observed a significant decrease in the number of newly internalized EBs in cells that contains a mature inclusion compared to uninfected cells (Figure 2G). This resistance to re-infection is partially mediated by CPAF, as treatment of pre-infected cells with anti-CPAF peptides increased EB entry (Figure 2H).
CPAF activity is required for Chlamydia replication and integrity of the inclusion
While CPAF has well-established functions in the down-regulation of immune responses, pro-apoptotic factors, and the processing of cytoskeletal proteins (Zhong, 2009), the role that these proteolytic events play in bacterial replication and intracellular survival is unknown. We treated Chlamydia-infected cells with anti-CPAF peptide and determined that peptide treatment significantly lowered yields of EBs and stunted inclusion growth (Figure 3A, Figure S3A–B). To assess the specificity of this peptide, we tested the effect of anti-CPAF peptide on HeLa cells infected with C. muridarum and C. caviae, two chlamydial species that display varying degrees of CPAF sequence conservation (Figure S3C). Consistent with the high conservation between C. trachomatis and C. muridarum CPAFs, the anti-CPAF peptide prevented cleavage of substrates by C. muridarum CPAF (Figure S3D) and blocked C. muridarum replication (Figure S3E). In contrast, and the divergent C. caviae CPAF was not sensitive to the anti-CPAF peptide in vitro or in vivo (Figure S3D, F).
Figure 3. CPAF is required for chlamydial replication and inclusion maintenance.
(A) Anti-CPAF peptide blocks the generation of Chlamydia infectious particles. L2-infected HeLa cells were treated at 12 hpi with increasing concentration of peptide. EBs were harvested at 30 hpi and Inclusion forming units (IFU) were quantified. Error bars represent +/− St. error.
(B–D) CPAF activity is required to maintain inclusion integrity. DNA was stained with Hoechst (blue). Inclusion integrity was assessed by immuno-localization of the inclusion membrane marker IncA (red) and Chlamydia (green) (B) and by transmission electron microscopy (C). Note collapse of IncA-positive membranes and loss of inclusion integrity with bacteria in the cytoplasm (arrows) of infected cells lacking CPAF activity (right panels). (D) Vimentin-positive inclusions were identified by IF of infected cells fixed 24 hpi subsequent to treatment with control and inhibitory peptide at 12 hpi. Error bars represent +/− St. error.
(E) Inhibition of CPAF activity leads to activation of inflammatory cytokines. Levels of IL-8 in supernatants from Chlamydia infected cells treated with inhibitory peptide as in (B–D) were determined by ELISA at 24 hpi. Error bars represent +/− St. error.
Next, we assessed the mechanism underlying the inhibition of Chlamydia replication by performing a microscopic analysis of infected cells treated with anti-CPAF peptide. Prolonged inhibition of CPAF (>8 h) with the anti-CPAF peptide, but not the scrambled control peptide, led to the collapse of the inclusion with the inclusion membrane markers localizing to perinuclear aggregates and dispersed bacteria (Figure 3B, Figure S4I). Analysis of these cells by transmission electron microscopy confirmed the loss of inclusion integrity and disruption of the inclusion membrane with intact Chlamydia freely residing in the cytoplasm (Figure 3C). Accordingly, we observed a loss of vimentin re-organization around the inclusion (Figure 3D), which has been previously linked to inclusion stability (Kumar and Valdivia, 2008). Consistent with the presence of intact bacteria in the cytoplasm, we also observed enhanced secretion of IL-8 (Figure 3E). Hence, we conclude that CPAF functions to maintain inclusion integrity during infection.
Inhibition of CPAF induces caspase-1-dependent cell death
In addition to limiting inclusion growth, treatment with anti-CPAF peptide led to a marked increase in the number of infected epithelial cells with condensed nuclei (Figure 4A, Figure S4A–B). This effect is specific to CPAF, as the anti-CPAF peptide did not lead to host cell death in C. caviae-infected cells (Figure S4E–F). Cell death occurred after 20 hpi (Figure 4A), which coincides with the translocation of CPAF into the host cytoplasm and was observed in several non-myeloid cell lines (Figure S4C–D). Besides resulting in a ruptured inclusion and cell death, inhibiting CPAF also led to the formation of multiple inclusions and small inclusions (Figure S4H). Given that Chlamydia-infected cells are highly resistant to intrinsic and extrinsic apoptotic stimuli (Greene et al., 2004) and that pro-apoptotic BH3-only proteins are CPAF targets (Paschen et al., 2008; Pirbhai et al., 2006), we hypothesized that CPAF inhibition led to the onset of apoptosis in infected cells. To test this, we labeled infected cells with propidium iodide and Annexin V to monitor cell viability and the loss of plasma membrane symmetry - a hallmark of apoptosis (Vermes et al., 1995). Chlamydia-infected cells treated with anti-CPAF peptide did not stain with Annexin V, indicating that the cell death is unlikely the result of classical apoptosis (Fig 4B). Consistent with this observation, we did not detect caspase-3 cleavage, another feature of apoptotic cell death (Figure 4C) (Galluzzi et al., 2009).
Figure 4. Inhibition of CPAF activity induces non-apoptotic death of host cells.

(A) Inhibiting CPAF activity induces host cell death. Inhibitory peptide was added to infected HeLa cells 12 hpi and the percentage of infected cells with condensed nuclei was quantified at the indicated times. Error bars represent +/− St. error.
(B–C) Inhibition of CPAF activity does not induce apoptosis. Infected cells were treated with inhibitory peptide and the percentage of AnnexinV-FITC positive cells among propidium iodide (PI) negative cells was determined by flow cytometry (B). In parallel samples, cleavage of caspase-3, a hallmark of apoptosome activation, was monitored by immunoblot analysis. Actin and MOMP are host and bacterial loading controls, respectively. Treatment with staurosporine (2 μM) and the pan-caspase inhibitor Ac-ZVAD-fmk were used as controls for the induction of apoptosis and caspase activity, respectively. Error bars represent +/− St. error.
(D) The pan-caspase inhibitor Ac-ZVAD-FMK blocks death of Chlamydia-infected epithelial cells. Infected HeLa cells were treated as in (A) in the presence or absence of Ac-ZVAD-fmk. Cell toxicity was determined by flow cytometry of PI-stained cells. PI staining was not seen in cells pre-treated with Ac-ZVAD-fmk or the scrambled control peptide. Error bars represent +/− St. error.
Unexpectedly, the pan-caspase inhibitor ZVAD-FMK efficiently blocked the death of infected cells treated with anti-CPAF peptide (Figure 4D). In myeloid cells, activation of caspase-1 by infectious agents can lead to pyroptosis, a cell death pathway accompanied by pore formation and the release of pro-inflammatory modifiers (Brennan and Cookson, 2000; Cervantes et al., 2008; Jesenberger et al., 2000). However, it is unclear whether this process also occurs in epithelial cells. We determined that the caspase-1 inhibitor Ac-YVAD-cmk efficiently blocked the death of infected epithelial cells treated with anti-CPAF peptide (Figure 5A). In addition, we observed that caspase-1 activity, as assessed with a fluorescently labeled caspase-1 substrate (Darzynkiewicz et al., 2011), was significantly increased during treatment with anti-CPAF peptide (Figure 5B). Next, we confirmed the role played by caspase-1 and its upstream activator, the inflammasome adaptor protein ASC (Martinon et al., 2002), in mediating cell death by infecting transformed mouse lung fibroblasts (MLF) derived from caspase-1 (ICE−/−) and ASC (Pycard−/−) knockout mice with C. trachomatis and C. muridarum. These MLF, unlike their wild-type counterparts, were resistant to host cell death in response to treatment with anti-CPAF peptide, indicating that inflammasome-dependent activation of caspase-1 is required for cell death (Figure 5C, Figure S4G). Interestingly, pharmacological or genetic inhibition of caspase-1 did not rescue bacterial replication (Figure 5D, Figure S3E). Overall, these findings indicate that CPAF plays at least two distinct roles in promoting chlamydial replication: mediating inclusion stability and suppressing caspase-1 dependent cell death.
Figure 5. Death of Chlamydia infected cells treated with CPAF inhibitory peptide is dependent on caspase-1.
(A) Caspase-1 inhibitors block Chlamydia-mediated cell death. Infected HeLa cells were treated with inhibitory peptide at 12 hpi, fixed at 24 hpi, and the percentage of cells with condensed nuclei was quantified. Ac-YVAD-cmk was added at 2 hpi. Error bars represent +/− St. error.
(B) Caspase-1 activation is enhanced during infection when CPAF is inhibited. Infected cells were labeled with a cell-permeable fluorescent substrate of caspase-1 and treated with anti-CPAF peptide, and caspase-1 activity at 28 h post infection was monitored by flow cytometry.
(C) Caspase-1 and ASC are required for Chlamydia-mediated host cell death. Chlamydia-infected lung fibroblasts derived from caspase-1 (ICE−/−) and ASC adaptor (ASC−/−) knockout mice were resistant to cell death induced by anti-CPAF peptide. Error bars represent +/− St. error.
(D) CPAF is required for chlamydial replication independently of its role in suppressing caspase-1 mediated cell death. Chlamydia replication and generation of IFUs in MLFs was assessed as in Fig 3. Note dose-dependent loss in IFU yields in all MLF lines. Error bars represent +/− St. error.
DISCUSSION
Chlamydia effector proteins, in a life-cycle stage-dependent manner, control bacterial entry, establishment of a replicative vacuole, modulation of innate immune responses and ultimately, exit from the host cell (Valdivia, 2008). The deployment of such a vast arsenal of bacterial proteins into the mammalian cytoplasm presents two challenges for Chlamydia: 1) how to limit the detection of these microbe-derived proteins by innate immune surveillance pathways and 2) how to control the function of these proteins after they leave the confines of the bacterial cell. Here we present evidence that CPAF, a chlamydial protease that modulates multiple host functions required for innate and adaptive immunity (Zhong, 2009), also cleaves a subset of effector proteins that are translocated early during infection. The concept of a “meta” effector, one that regulates the function of others, was proposed for the Legionella E3 ubiquitin ligase LubX, which ubiquitinates the effector SidH and targets it for degradation (Kubori, 2010). Our data suggests that CPAF serves a similar function by directly cleaving a subset of secreted effectors and inclusion membrane proteins.
Because CPAF is translocated into the cytoplasm starting at 16 h post-infection (Shaw et al., 2002), the inclusion membrane proteins we identified as CPAF-substrates are unlikely to be cleaved until the middle stage of the infectious cycle. This is consistent with the observation that the steady-state levels of Ct005, IncC and IncD do not decrease until later in infection (Figure 1D). CPAF cleavage of Incs during mid cycle may be less efficient due to Inc-modification or formation of Inc complexes that mask CPAF recognition sites. In addition, substrate specific features (e.g. secondary or tertiary structure) may influence the efficiency of CPAF-mediated processing. Among the 42 putative Inc proteins (Li et al., 2008) tested, only four were identified as CPAF substrates (Figure 1), and three of these are predicted to be expressed within the first 3 h of infection (Belland et al., 2003). Notably, not all Inc proteins were targets of degradation, suggesting that CPAF strategically removes a subset of Inc proteins rather than achieving a wholesale remodeling of the inclusion membrane. Inc proteins are predicted to perform multiple functions including protection from apoptosis (Verbeke et al., 2006), sequestration of signaling proteins (Mital et al., 2010), and re-routing of membrane transport (Rzomp et al., 2006). We postulate that factors required early in the biogenesis of the inclusion may not be necessary for – or may even be detrimental to – optimal inclusion expansion later in the infectious cycle. As such, cleavage of these early Inc proteins may be essential to the chlamydial life cycle. It is also possible that CPAF cleavage of Inc-proteins may limit their detection by microbial pattern recognition receptors or their processing and display by MHC class I molecules (Roan and Starnbach, 2008). Given CPAF degradation of transcription factors required for MHC-I and MHC-II expression (Zhong et al., 2001), cleavage of Inc proteins could further reduce the possibility of these proteins being presented as antigens.
Two chlamydial CPAF targets (Tarp and Ct694) are translocated during EB attachment. One condition under which EB effectors would encounter CPAF in the host cytoplasm is if an EB where to infect a cell that already had an established inclusion. In such a scenario, CPAF would play a role in preventing the establishment of secondary inclusions that may impair development of the primary inclusion. Consistent with this, Tarp translocated by EBs into cells with established inclusions is rapidly degraded (Figure 2). This degradation is partially blocked upon delivery of a peptide-based CPAF inhibitor, indicating that Tarp is a true target of CPAF-mediated degradation. Accordingly, EBs are blocked from entering a cell that contains a mature inclusion, but this is reversed when CPAF activity is inhibited. Collectively, CPAF may therefore mediate resistance to superinfection by degrading secreted effectors involved in invasion and early inclusion biogenesis.
CPAF-specific inhibitors revealed that this protease plays an essential role in chlamydial development and replication (Figure 3, Figure S4–5). Inclusion growth was stunted concomitantly with a drop in the generation of infectious progeny and marked increase in toxicity to the infected cell (Figure 3–4). The most striking effect of anti-CPAF peptide was the loss of inclusion integrity in treated cells. Whether this is the consequence of a lack of Inc protein turnover or processing of intermediate filaments at the inclusion membrane is unclear. Regardless of the sequence of events leading to inclusion membrane dismantling, it is apparent that this protease plays an essential role in maintaining the integrity of the pathogenic vacuole.
CPAF is predicted to both prevent (Pirbhai et al., 2006) and promote (Paschen et al., 2008) cell death depending on the stage of infection. The pro-survival mechanism of CPAF may include the degradation of pro-apoptotic proteins, and indeed, we found that anti-CPAF peptide blocked the cleavage of Puma (Figure 2C). However, Chlamydia-infected cells treated with anti-CPAF peptide initiated a cell death program that did not resemble classical apoptosis. Instead, we found that death of infected epithelial cells upon inhibition of CPAF activity was dependent on caspase-1 and that anti-CPAF inhibitors led to an early activation of caspase-1 (Figure 3, 5). In myeloid cells, caspase-1 dependent cell death – also termed pyroptosis – results in rapid plasma membrane rupture and release of pro-inflammatory cytokines such as IL-1β and IL-18 (Bergsbaken et al., 2009). Pore formation in the plasma membrane disrupts ionic gradients, which increases the osmotic pressure leading to swelling and lysis. Normally, caspase-1-mediated cell death aids in the clearance of infections (Bergsbaken and Cookson, 2007; Brennan and Cookson, 2000; Sansonetti et al., 2000). Recently, it has been reported that Chlamydia infection activates caspase-1 in epithelial cells late during infection and is required for optimal chlamydial growth (Abdul-Sater et al., 2009; Lu et al., 2000). Potentially, the dependence of chlamydial growth on caspase-1 is a consequence of enhanced lipid biosynthesis that occurs upon activation of low levels of caspase-1 (Gurcel et al., 2006). However, upon inhibition of CPAF, caspase-1 activation occurs much earlier and with greater magnitude, most likely as a result of an overwhelming response to bacterial products in the cytoplasm. We predict that this premature activation of caspase-1 triggers an irreversible cell death program in Chlamydia-infected non-myeloid cells, as has been recently proposed in Salmonella that resides in the cytoplasm of infected colonic epithelial cells (Knodler et al., 2010). It is also possible that CPAF plays a more direct role in suppressing inflammasome formation as has been demonstrated for the Yersinia effectors YopE and YopT (Schotte et al., 2004) and the poxvirus pyrin-domain protein M13L (Johnston et al., 2005).
Overall, these studies establish that CPAF is an essential Chlamydia virulence factor with an important role in maintaining the integrity of the inclusion and preventing the premature death of its host cell in a single-cell, autonomous tissue culture model of infection (Figure 6). Furthermore, CPAF plays a previously unappreciated role in the turnover of other effectors to aid in the remodeling of the inclusion and to limit secondary infections. Consistent with its obligate intracellular lifestyle and its small genome, it is not surprising that Chlamydia has become adept at multi-tasking by expressing regulatory effectors with multiple functions, like CPAF. Thus, CPAF emerges as a central virulence protein and an attractive target for the design of specific protease inhibitors for therapeutic intervention.
Figure 6. CPAF mediates resistance to re-infection, inclusion integrity and protection from caspase-1-mediated cell death.
CPAF reorganizes intermediate filaments at the inclusion periphery to promote inclusion stability (Kumar and Valdivia, 2008) and mediates protection from re-infection by targeting effectors translocated during invasion and early inclusion biogenesis. Inhibition of CPAF activity leads to a loss of protection from re-infection. In addition, CPAF activity is required to maintain inclusion membrane integrity, dampen the secretion of pro-inflammatory cytokines, and prevent inflammasome-dependent activation of caspase-1-dependent cell death.
EXPERIMENTAL PROCEDURES
Expression constructs and proteolysis screen
C. trachomatis ORFs cloned into the yeast expression vector pSDY8 (C-terminal GFP tag) (Sisko et al., 2006) or the E. coli expression vector pGEX-4T-1 vector (GE Healthcare) are listed in Table S1. Hexahistidine-tagged CPAF (pET30b) was purified as previously described (Huang et al., 2008). For in vitro cleavage assays, Chlamydia proteins fused to EGFP or GST were expressed in yeast and E. coli respectively. Yeast lysates or purified GST-tagged were incubated with cytosols from uninfected or LGV-L2 infected (40 h) HeLa cells for 30 min at 37°C. Processed chlamydial proteins were detected by immunoblot analysis with anti-GFP antibodies or Coomassie Blue staining.
Cell culture and Chlamydia infections
MLFs from Pycard−/−, ICE−/− and WT mice were isolated as previously described (van Deventer et al., 2008). Ex vivo lungs were minced, incubated with 1 mg/ml collagenase A and 20μg/ml DNAse I in RPMI supplemented with 2% fetal calf serum (FBS) for 45 min at 37°C. Digested lungs were filtered, washed and red blood cells were lysed in NH4CaK buffer. Cultured MLFs were immortalized by lentiviral mediated transduction of T-antigen and human telomerase as previously described (O’Hayer and Counter, 2006). HeLa cells (ATCC) and MLFs were maintained in DMEM supplemented with 10% FBS (CellGro Mediatech Inc). C. trachomatis LGV-L2 434/Bu was propagated in HeLa cells as previously described (Caldwell et al., 1981). EBs were added to HeLa cells at the indicated MOIs and infections were synchronized by centrifugation at 300×g for 30 min at 4ºC. C. muridarum Nigg and C. caviae stocks were kind gifts from Roger Rank (University of Arkansas).
Generation of antibodies and immunodetection protocols
Rabbits were immunized with recombinant GST fusions to Ct005, IncC, IncD, Tarp (Ted Hackstadt, RML/NIH) and hexahistidine-tagged CPAF (J. Chai, Institute of Biological Sciences, Beijing) produced in E. coli BL21-DE3 (Stratagene) and purified by affinity chromatography. IgG antibodies were purified with Protein A-coated Sepharose beads (GE Healthcare). Membrane-associated chlamydial proteins were harvested from infected HeLa by ultracentrifugation of whole cell lysates on an Optiprep (Sigma) discontinuous density gradient (25, 20, 17.5, 15, 12.5, 10%) and assessing the fractionation of IncA and IncG positive membranes by immunoblot analysis. To assess CPAF cleavage of membrane proteins and EB proteins, purified membranes and soluble EB protein lysate were incubated with 6xHis-CPAF at 37°C for 20 min, and resulting product analyzed by immunoblots with specific antibodies.
Generation of Inhibitory peptide and CPAF cleavage assays
The anti-CPAF peptide (SLFYSPMVPHFWAELRNHYATSGLKRRRRRRRRR) and scrambled control (NFALSHFRLPLSTYKEMPYVSHWAGRRRRRRRRR) peptide were synthesized by solid-phase peptide synthesis by the FMOC t-Boc method. Peptides were purified to homogeneity by reverse-phase HPLC, and confirmed by mass spectrometry. Scaled up production of peptides were performed by Eton Biosciences (Durham, NC) and purified by HPLC. IC50 values were determined by assessing cleavage inhibition of an Abz-tagged Vimentin peptide VRLRSSVPGV-NH2. His-CPAF was pre-incubated with 10μM Lactacystin (EMD), solvent control (DMSO), ACP or CP at 37ºC for 30 min. Cleavage products were assessed by HPL analysis. To assess CPAF-dependent cleavage during infection, 1.2×106 HeLa cells were grown in 6-well plates and infected with LGV-L2 at an MOI of 1, treated with anti-CPAF or control peptide at 12 hpi and harvested at 30 hpi.
Immunofluorescence microscopy (IF)
A detailed list of antibodies used in this study is available in supplemental data (Table S3). For routine indirect immunofluorescence, 0.2×106 HeLa cells were grown on glass coverslips and infected at the indicated MOI. Cells were fixed with cold 3% formaldehyde in phosphate-buffered saline (PBS), permeabilized with 0.1% Triton X-100, blocked in 2% bovine serum albumin (BSA) and incubated with the specified primary antibodies in 2%BSA in PBS and fluorophore-conjugated anti-rabbit or anti-mouse goat IgG (Molecular Probes). Host and bacterial DNA were stained with 1μM Hoechst in PBS (Invitrogen). Infected cells were imaged with a Zeiss Axioscope epifluorescence microscope and Axiovision v3.0 software or a Leica TCS SL confocal microscope and processed with Leica software. For transmission electron microscopy (TEM), HeLa cells grown on thermanox (Electron Microscopy Services) coverslips were infected with LGV-L2 and treated with anti-CPAF peptide as described above, fixed with 0.05% malachite green/2.5% gluteraldehyde, postfixed with 0.8% osmium tetroxide and 1% tannic acid and 1% uranyl acetate. Following dehydration with ethanol of samples, sections were post-stained and imaged with a Tecnai G12 Twin electron microscope (FEI).
Radiolabeling and secondary infections
HeLa cells were infected with C. trachomatis for 18 h and labeled with 300μCi S35 cysteine/methionine (Perkin Elmer) in the presence of 40μg/ml cyclohexamide (Sigma) for an additional 22 h. EB seed was harvested following gentle sonication and stored at −80°C in SPG buffer (0.25M sucrose, 10 mM sodium phosphate, 5 mM L-glutamic acid). Uninfected or HeLa cells that had been pre-infected for 30 h were infected with unlabeled or S35-labeled LGV-L2 seed at an MOI of 50. Cells were washed extensively with trypsin, and harvested at 10 min post the secondary infection in lysis buffer (20 mM Tris, 150 mM NaCl, 1% Tx100, 2 mM PMSF, 2 mM MG132, 10 mM ALLN, protease inhibitor cocktail (Roche)). Tarp and MOMP were immunoprecipitated using anti-TARP and anti-MOMP polyclonal antisera and Protein A-coated Sepharose beads (GE Healthcare) and detected in a Typhoon9410 Variable Image PhosphorImager (Amersham Biosciences), and quantified using ImageQuant 5.1TL (GE Healthcare). To test the effect of inhibitory peptide, cells were treated with 12μM peptides for the duration of the secondary infections. To distinguish intracellular from extracellular EBs, cells were infected with CellTracker (Invitrogen)-labeled EBs for 30 min, fixed without premeabilization and extracellular EBs were immunostained with an anti-L2 antisera.
Cytokine, apoptosis, and caspase-1 activation assays
HeLa cells were infected with C. trachomatis LGV-L2 at an MOI of 1. At 3 hpi, cells were treated with 40μM Z-VAD-FMK (Promega) or 400μM Ac-YVAD-cmk (Enzo Life Sciences). At 12 hpi, cells were treated with 12.5μM of anti-CPAF peptide or control peptide. IL-8 secretion into the media was determined with a Human IL-8 ELISA kit (BioLegend) as recommended by the manufacturer. Apoptotic cells were identified with an AnnexinV-FLOUS Staining Kit (Roche) and activation of caspase-1 was determined by labeling active caspase-1 with a Carboxyfluorescein FLICA Detection Kit (Immunochemistry) and analyzed in a FACScanner (BD).
Supplementary Material
Acknowledgments
We thank J.D. Dunn, J. Cocchiaro and J. Coers for helpful comments on the manuscript, E. Snavely for technical assistance, A. Xavier, Y. Chen, R. Rank, J. Chai and T. Hackstadt for generous gifts of cell lines, strains, antibodies and expression plasmids, and R. Flavell and V. Dixit for knockout mice. This work was supported by NIH awards AI081694 (RHV), AI46611 (DM) and U54 AI057157 (J. PY. T), the Burroughs Wellcome Trust Program in the Pathogenesis of Infectious Diseases (RHV), the Crohn’s and Colitis Foundation of America (BD) and a CR Hauser Fellowship (MB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdul-Sater AA, Koo E, Hacker G, Ojcius DM. Inflammasome-dependent caspase-1 activation in cervical epithelial cells stimulates growth of the intracellular pathogen Chlamydia trachomatis. J Biol Chem. 2009;284:26789–26796. doi: 10.1074/jbc.M109.026823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adderley-Kelly B, Stephens EM. Chlamydia: A major health threat to adolescents and young adults. ABNF J. 2005;16:52–55. [PubMed] [Google Scholar]
- Arnold R, Brandmaier S, Kleine F, Tischler P, Heinz E, Behrens S, Niinikoski A, Mewes HW, Horn M, Rattei T. Sequence-based prediction of type III secreted proteins. PLoS Pathog. 2009;5:e1000376. doi: 10.1371/journal.ppat.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J, Beatty WL, Caldwell HD. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci U S A. 2003;100:8478–8483. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsbaken T, Cookson BT. Macrophage activation redirects yersinia-infected host cell death from apoptosis to caspase-1-dependent pyroptosis. PLoS Pathog. 2007;3:e161. doi: 10.1371/journal.ppat.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts HJ, Wolf K, Fields KA. Effector protein modulation of host cells: examples in the Chlamydia spp. arsenal. Curr Opin Microbiol. 2009;12:81–87. doi: 10.1016/j.mib.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Brennan MA, Cookson BT. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol Microbiol. 2000;38:31–40. doi: 10.1046/j.1365-2958.2000.02103.x. [DOI] [PubMed] [Google Scholar]
- Caldwell HD, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes J, Nagata T, Uchijima M, Shibata K, Koide Y. Intracytosolic Listeria monocytogenes induces cell death through caspase-1 activation in murine macrophages. Cell Microbiol. 2008;10:41–52. doi: 10.1111/j.1462-5822.2007.01012.x. [DOI] [PubMed] [Google Scholar]
- Chen D, Lei L, Lu C, Flores R, DeLisa MP, Roberts TC, Romesberg FE, Zhong G. Secretion of the chlamydial virulence factor CPAF requires the Sec-dependent pathway. Microbiology. 2010;156:3031–3040. doi: 10.1099/mic.0.040527-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian JG, Vier J, Paschen SA, Hacker G. Cleavage of the NF-{kappa}B-family protein p65/RelA by the chlamydial protease chlamydial protease-like activity factor (CPAF) impairs pro-inflammatory signalling in cells Infected with chlamydiae. J Biol Chem. 2010 doi: 10.1074/jbc.M110.152280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci U S A. 2004;101:10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Pozarowski P, Lee BW, Johnson GL. Fluorochrome-labeled inhibitors of caspases: convenient in vitro and in vivo markers of apoptotic cells for cytometric analysis. Methods Mol Biol. 2011;682:103–114. doi: 10.1007/978-1-60327-409-8_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Su H, Huang Y, Zhong Y, Zhong G. Cleavage of host keratin 8 by a Chlamydia-secreted protease. Infect Immun. 2004;72:3863–3868. doi: 10.1128/IAI.72.7.3863-3868.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Aaronson SA, Abrams J, Alnemri ES, Andrews DW, Baehrecke EH, Bazan NG, Blagosklonny MV, Blomgren K, Borner C, et al. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ. 2009;16:1093–1107. doi: 10.1038/cdd.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene W, Xiao Y, Huang Y, McClarty G, Zhong G. Chlamydia-infected cells continue to undergo mitosis and resist induction of apoptosis. Infect Immun. 2004;72:451–460. doi: 10.1128/IAI.72.1.451-460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurcel L, Abrami L, Girardin S, Tschopp J, van der Goot FG. Caspase-1 activation of lipid metabolic pathways in response to bacterial pore-forming toxins promotes cell survival. Cell. 2006;126:1135–1145. doi: 10.1016/j.cell.2006.07.033. [DOI] [PubMed] [Google Scholar]
- Hower S, Wolf K, Fields KA. Evidence that CT694 is a novel Chlamydia trachomatis T3S substrate capable of functioning during invasion or early cycle development. Mol Microbiol. 2009;72:1423–1437. doi: 10.1111/j.1365-2958.2009.06732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu VH, Harding-Esch EM, Burton MJ, Bailey RL, Kadimpeul J, Mabey DCW. Tropical Medicine & International Health. 2010. Epidemiology and control of trachoma: systematic review; pp. 673–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Feng Y, Chen D, Wu X, Huang S, Wang X, Xiao X, Li W, Huang N, Gu L, et al. Structural basis for activation and inhibition of the secreted chlamydia protease CPAF. Cell Host Microbe. 2008;4:529–542. doi: 10.1016/j.chom.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Jesenberger V, Procyk KJ, Yuan J, Reipert S, Baccarini M. Salmonella-induced caspase-2 activation in macrophages: a novel mechanism in pathogen-mediated apoptosis. J Exp Med. 2000;192:1035–1046. doi: 10.1084/jem.192.7.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett TJ, Dooley CA, Mead DJ, Hackstadt T. Chlamydia trachomatis tarp is phosphorylated by src family tyrosine kinases. Biochem Biophys Res Commun. 2008;371:339–344. doi: 10.1016/j.bbrc.2008.04.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JB, Barrett JW, Nazarian SH, Goodwin M, Ricciuto D, Wang G, McFadden G. A poxvirus-encoded pyrin domain protein interacts with ASC-1 to inhibit host inflammatory and apoptotic responses to infection. Immunity. 2005;23:587–598. doi: 10.1016/j.immuni.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Kawana K, Quayle AJ, Ficarra M, Ibana JA, Shen L, Kawana Y, Yang H, Marrero L, Yavagal S, Greene SJ, et al. CD1d degradation in Chlamydia trachomatis-infected epithelial cells is the result of both cellular and chlamydial proteasomal activity. J Biol Chem. 2007;282:7368–7375. doi: 10.1074/jbc.M610754200. [DOI] [PubMed] [Google Scholar]
- Knodler LA, Vallance BA, Celli J, Winfree S, Hansen B, Montero M, Steele-Mortimer O. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc Natl Acad Sci U S A. 2010;107:17733–17738. doi: 10.1073/pnas.1006098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T, Shinzawa N, Kanuka H, Nagai H. Legionella Metaeffector Exploits Host Proteasome to Temporally Regulate Cognate Effector. PLoS Pathog. 2010:6. doi: 10.1371/journal.ppat.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar Y, Valdivia RH. Actin and intermediate filaments stabilize the Chlamydia trachomatis vacuole by forming dynamic structural scaffolds. Cell Host Microbe. 2008;4:159–169. doi: 10.1016/j.chom.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chen C, Chen D, Wu Y, Zhong Y, Zhong G. Characterization of fifty putative inclusion membrane proteins encoded in the Chlamydia trachomatis genome. Infect Immun. 2008;76:2746–2757. doi: 10.1128/IAI.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Shen C, Brunham RC. Chlamydia trachomatis infection of epithelial cells induces the activation of caspase-1 and release of mature IL-18. J Immunol. 2000;165:1463–1469. doi: 10.4049/jimmunol.165.3.1463. [DOI] [PubMed] [Google Scholar]
- Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- Mehlitz A, Banhart S, Hess S, Selbach M, Meyer TF. Complex kinase requirements for Chlamydia trachomatis Tarp phosphorylation. FEMS Microbiol Lett. 2008;289:233–240. doi: 10.1111/j.1574-6968.2008.01390.x. [DOI] [PubMed] [Google Scholar]
- Mital J, Miller NJ, Fischer ER, Hackstadt T. Specific chlamydial inclusion membrane proteins associate with active Src family kinases in microdomains that interact with the host microtubule network. Cell Microbiol. 2010;12:1235–1249. doi: 10.1111/j.1462-5822.2010.01465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hayer KM, Counter CM. A genetically defined normal human somatic cell system to study ras oncogenesis in vivo and in vitro. Methods Enzymol. 2006;407:637–647. doi: 10.1016/S0076-6879(05)07050-3. [DOI] [PubMed] [Google Scholar]
- Paschen SA, Christian JG, Vier J, Schmidt F, Walch A, Ojcius DM, Hacker G. Cytopathicity of Chlamydia is largely reproduced by expression of a single chlamydial protease. J Cell Biol. 2008;182:117–127. doi: 10.1083/jcb.200804023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirbhai M, Dong F, Zhong Y, Pan KZ, Zhong G. The secreted protease factor CPAF is responsible for degrading pro-apoptotic BH3-only proteins in Chlamydia trachomatis-infected cells. J Biol Chem. 2006;281:31495–31501. doi: 10.1074/jbc.M602796200. [DOI] [PubMed] [Google Scholar]
- Roan NR, Starnbach MN. Immune-mediated control of Chlamydia infection. Cell Microbiol. 2008;10:9–19. doi: 10.1111/j.1462-5822.2007.01069.x. [DOI] [PubMed] [Google Scholar]
- Rzomp KA, Moorhead AR, Scidmore MA. The GTPase Rab4 interacts with Chlamydia trachomatis inclusion membrane protein CT229. Infect Immun. 2006;74:5362–5373. doi: 10.1128/IAI.00539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonetti PJ, Phalipon A, Arondel J, Thirumalai K, Banerjee S, Akira S, Takeda K, Zychlinsky A. Caspase-1 activation of IL-1beta and IL-18 are essential for Shigella flexneri-induced inflammation. Immunity. 2000;12:581–590. doi: 10.1016/s1074-7613(00)80209-5. [DOI] [PubMed] [Google Scholar]
- Schotte P, Denecker G, Van Den Broeke A, Vandenabeele P, Cornelis GR, Beyaert R. Targeting Rac1 by the Yersinia effector protein YopE inhibits caspase-1-mediated maturation and release of interleukin-1beta. J Biol Chem. 2004;279:25134–25142. doi: 10.1074/jbc.M401245200. [DOI] [PubMed] [Google Scholar]
- Scidmore-Carlson MA, Shaw EI, Dooley CA, Fischer ER, Hackstadt T. Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol Microbiol. 1999;33:753–765. doi: 10.1046/j.1365-2958.1999.01523.x. [DOI] [PubMed] [Google Scholar]
- Shaw AC, Vandahl BB, Larsen MR, Roepstorff P, Gevaert K, Vandekerckhove J, Christiansen G, Birkelund S. Characterization of a secreted Chlamydia protease. Cell Microbiol. 2002;4:411–424. doi: 10.1046/j.1462-5822.2002.00200.x. [DOI] [PubMed] [Google Scholar]
- Sisko JL, Spaeth K, Kumar Y, Valdivia RH. Multifunctional analysis of Chlamydia-specific genes in a yeast expression system. Mol Microbiol. 2006;60:51–66. doi: 10.1111/j.1365-2958.2006.05074.x. [DOI] [PubMed] [Google Scholar]
- Stewart KM, Horton KL, Kelley SO. Cell-penetrating peptides as delivery vehicles for biology and medicine. Org Biomol Chem. 2008;6:2242–2255. doi: 10.1039/b719950c. [DOI] [PubMed] [Google Scholar]
- Sun J, Kintner J, Schoborg RV. The host adherens junction molecule nectin-1 is downregulated in Chlamydia trachomatis-infected genital epithelial cells. Microbiology. 2008;154:1290–1299. doi: 10.1099/mic.0.2007/015164-0. [DOI] [PubMed] [Google Scholar]
- Valdivia RH. Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr Opin Microbiol. 2008;11:53–59. doi: 10.1016/j.mib.2008.01.003. [DOI] [PubMed] [Google Scholar]
- van Deventer HW, Wu QP, Bergstralh DT, Davis BK, O’Connor BP, Ting JP, Serody JS. C-C chemokine receptor 5 on pulmonary fibrocytes facilitates migration and promotes metastasis via matrix metalloproteinase 9. Am J Pathol. 2008;173:253–264. doi: 10.2353/ajpath.2008.070732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeke P, Welter-Stahl L, Ying S, Hansen J, Hacker G, Darville T, Ojcius DM. Recruitment of BAD by the Chlamydia trachomatis vacuole correlates with host-cell survival. PLoS Pathog. 2006;2:e45. doi: 10.1371/journal.ppat.0020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- Yu H, Schwarzer K, Forster M, Kniemeyer O, Forsbach-Birk V, Straube E, Rodel J. Role of high-mobility group box 1 protein and poly(ADP-ribose) polymerase 1 degradation in Chlamydia trachomatis-induced cytopathicity. Infect Immun. 2010;78:3288–3297. doi: 10.1128/IAI.01404-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G. Killing me softly: chlamydial use of proteolysis for evading host defenses. Trends Microbiol. 2009;17:467–474. doi: 10.1016/j.tim.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G, Fan P, Ji H, Dong F, Huang Y. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J Exp Med. 2001;193:935–942. doi: 10.1084/jem.193.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G, Liu L, Fan T, Fan P, Ji H. Degradation of transcription factor RFX5 during the inhibition of both constitutive and interferon gamma-inducible major histocompatibility complex class I expression in chlamydia-infected cells. J Exp Med. 2000;191:1525–1534. doi: 10.1084/jem.191.9.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.