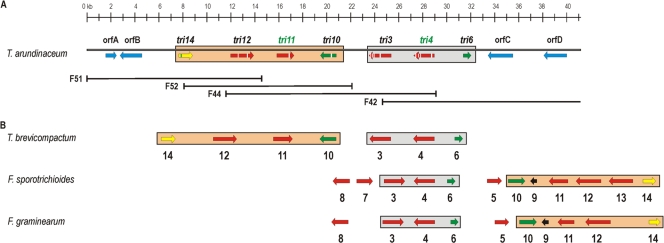
Fig. 1.
(A) Genomic organization of the Trichoderma arundinaceum IBT 40837 HA core cluster of tri genes. Overlapping phages representing the genomic region comprising the trichothecene biosynthetic gene cluster are also represented. The arrows indicate predicted positions and directions of transcription of individual genes/open reading frames (ORFs). Small rectangles inside each gene indicate the introns of each ORF. The names above each arrow correspond with their Fusarium orthologue designation. The red arrows refer to those structural genes involved in the HA biosynthesis (tri11, tri3, tri4) or in the export of HA outside the cell (tri12), the green arrows refer to those genes with regulatory functions in the trichothecene biosynthesis (tri6 and tri10), the yellow arrow (tri14) has an unknown function, and the blue arrows indicate non-tri genes. The genes tri11 and tri4, whose names are illustrated in green, were those whose role in the biosynthesis have been studied in the present work by expression in yeast and, in the case of tri4, also in F. verticillioides. Note that the tri5 gene has not been illustrated, even when it has been also expressed in yeast, since it is located in another region of the Ta37 genome outside the core cluster of tri genes. (B) Genomic organization of the trichothecene gene cluster of T. arundinaceum in comparison with T. brevicompactum and Fusarium species (10). The arrows indicate predicted positions and directions of transcription of individual genes/ORFs. Numbers indicate gene designations based on their Fusarium orthologues (e.g., 6 refers to tri6 or TRI6). Colored designations illustrate their role in trichothecene biosynthesis, as indicated above. Colored rectangles represent those genes that are grouped in a similar fashion in the different clusters.