Fig. 3.
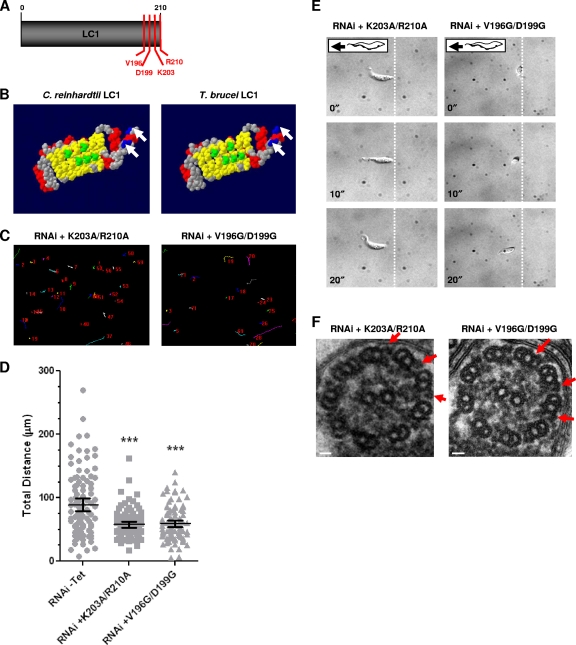
Identification of LC1 mutants with loss-of-function point mutations in procyclic cells. (A) LC1 schematic showing residues selected for mutational analysis (V196, D199, K203, and R210). (B) Space-filling model of C. reinhardtii LC1 (48) compared with predicted T. brucei LC1 (3). C. reinhardtii LC1 residues that are predicted to bind to the outer dynein γ heavy-chain (green) and basic residues in the C terminus (blue, arrows) are conserved in the T. brucei protein. Red, α helices; yellow, β sheets; gray, residues with no functional predictions. Reprinted from reference 3, with permission. (C) Motility traces of LC1 mutants with K203A/R210A and V196G/D199G point mutations in procyclic cells, demonstrating motility defects in both mutants. Cells were incubated in the presence or absence of tetracycline, and motility traces were carried out as described in the legend to Fig. 2C. (D) Quantification of total distance traveled by individual cells in motility traces was carried out as described in the legend to Fig. 2D. Horizontal lines indicate the mean of each data set, with bars indicating the 95% confidence interval. Results for the data sets were compared to those for RNAi without Tet. ***, significant difference (P < 0.0001). (E) Time-lapse images taken from Videos S5 and S6 in the supplemental material demonstrate that the K203A/R210A and V196G/D199G mutants do not move backward. Dashed lines indicate the starting position of the posterior end of the cell, and time stamps (in seconds) are indicated. (F) Transmission electron microscopy analysis of flagellum ultrastructure in procyclic LC1 mutants with point mutations (K203A/R210A and V196G/D199G) shows that outer dynein arms are present (arrows). Images are oriented with the PFR at the bottom of the frame. Bars, 30 nm.