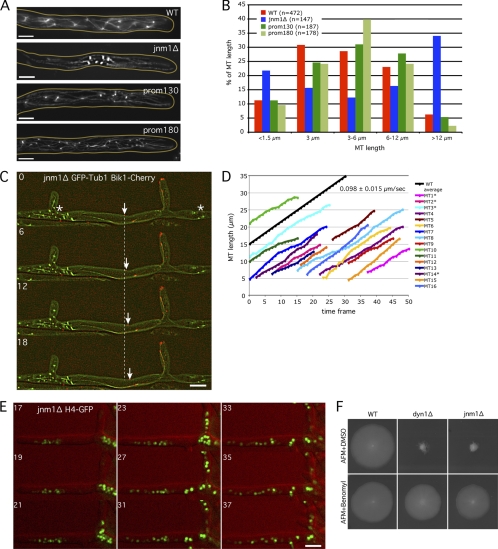
Fig. 7.
The very long cMTs observed in dynein/dynactin mutants are responsible for nuclear clustering. (A) Anti-alpha-tubulin immunostaining of jnm1Δ, prom130-DYN1, prom180-DYN1, and wild-type strains. Pictures correspond to stacks of 5 Z planes with 0.75-μm distance. (B) Quantification of cMT length in WT and mutant cells using anti-Tub1 immunostaining pictures, as shown in panel A. (C) Localization of Bik1-Cherry (red) at cMTs +ends (GFP-Tub1 in green) in jnm1Δ cell. Nuclei/SPBs cluster at some places (*), while long cMTs investigate the nuclei-free lateral branch. The arrow points to one growing cMT +end. The dashed line represents the position of this +end at time point 0. Time is indicated in seconds. (D) Quantification of the cMT polymerization rate using 1-Z-plane movies as shown in panel C. cMTs were measured from their –end (SPB observed with GFP-Tub1) to their +tip (Bik1-Cherry signal). Due to increased cMT length in the jnm1Δ mutant, SPBs and +tips were often in different focal planes (*), corresponding to the real length of the cMTs (SPB to +tip), whereas the other cMTs have an underestimated length (visible part of the cMT to the +tip). One frame corresponds to 6 s. The average cMT polymerization rate in the wild type is shown in black as a reference (see reference 8). (E) Nuclear clusters disappear and nuclei redistribute after treatment of jnm1Δ H4-GFP cells with 15 μg/ml nocodazole (red, DIC; green, H4-GFP). Time indicates minutes after addition of nocodazole. (F) Growth defects of jnm1Δ and dyn1Δ cells are partially rescued by the MT-destabilizing drug benomyl. Bars, 5 μm.