Fig. 1.
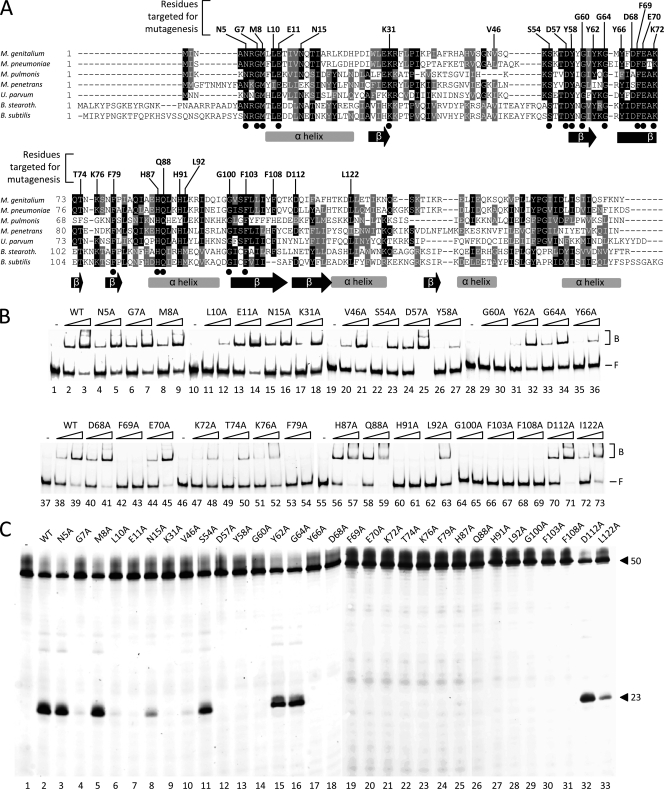
Multiple alignment of RecU sequences and activities of single point mutants of RecUMge. (A) A multiple alignment was generated with amino acid sequences predicted to be encoded by the following ORFs (with GenBank accession numbers in parentheses), M. genitalium G37 MG352 (Q49422-1), M. pneumoniae MAC MPN528a, Mycoplasma pulmonis recU (Q98RC3), Mycoplasma penetrans recU (Q8EWG0), Ureaplasma parvum recU (Q9PQJ4), B. stearothermophilus recU (Q5KXY4), and B. subtilis recU (P39792-1). The amino acid residues indicated above the sequences represent the RecUMge residues that were targeted by site-directed mutagenesis. The dots below the sequences indicate residues that are completely conserved among the RecU sequences. The gray bars and black arrows below the sequences represent α-helices and β-sheets, respectively, as determined previously for the RecUBsu protein (18). The multiple alignment was performed using Clustal W (http://www.ebi.ac.uk/Tools/clustalw/index.html). The program BOXSHADE 3.21 (http://www.ch.embnet.org/software/BOX_form.html) was used to generate white letters on black boxes (for residues that are identical in at least four out of seven sequences) and white letters on gray boxes (for similar residues). (B) HJ DNA-binding activities of point mutants of RecUMge. The binding of the proteins to the FAM-labeled HJ substrate HJ 1.1 was performed as indicated in Materials and Methods. In short, reactions were performed in volumes of 10 μl and contained 12.3 nM DNA substrate and either 0 nM protein (the lanes marked −) or two different concentrations of protein (100 nM or 400 nM), as indicated above the lanes. The protein-DNA mixtures were electrophoresed through native 6% polyacrylamide gels and visualized by fluorometry. The positions of the free, unbound DNA substrates in the gels are indicated by F on the right; the protein-bound substrates are indicated by B. (C) Holliday junction resolution by point mutants of RecUMge. The resolution reactions were performed as described in Materials and Methods, using the FAM-labeled HJ substrate HJ 1.1 and 400 nM protein. After the reaction (15 min at 37°C), the samples were analyzed by denaturing 15% polyacrylamide gel electrophoresis, followed by fluorometry. The lengths of the FAM-labeled strands from the DNA substrate (HJ12; 50 nucleotides) and the specific, major product from cleavage of the substrate by the mutants of RecUMge (23 nucleotides [26]) are indicated on the right. The names of the mutant proteins are shown above the lanes.