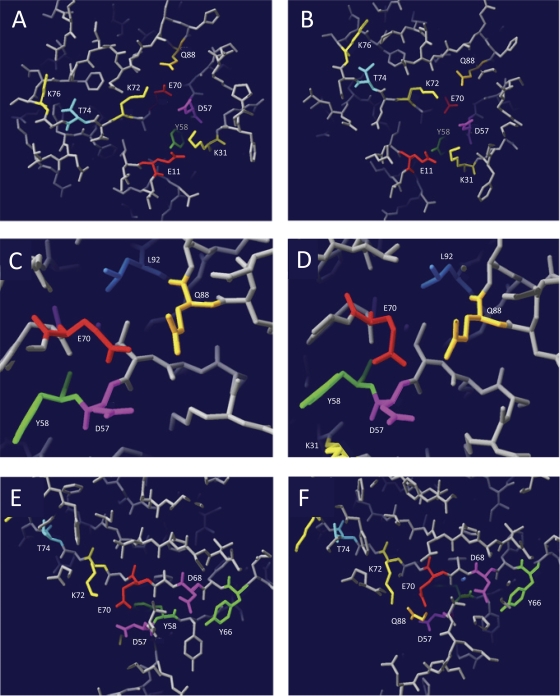
Fig. 2.
Three-dimensional models of the putative catalytic site of RecUMge. Three-dimensional models of RecUMge were constructed using the protein homology/analogy recognition engine Phyre (16). The lowest Phyre E values (3.4e−22 and 3e−21), with 100% estimated precision, were obtained with the structures of RecUBst and RecUBsu, respectively. The generated structure (PDB) files, which were based on either the RecUBst structure (A, C, and E) or the RecUBsu structure (B, D, and F), were analyzed using DeepView/Swiss-PdbViewer v4.0.1 (11). (A and B) Overview of the putative catalytic site of RecUMge. The group C amino acid residues, i.e., the amino acids that are crucial in the catalysis of HJ resolution by RecUMge but are not essential for HJ binding (Tables 1 and 2), are indicated by their names and residue-specific colors. In this slab view of the structure, the positions of 9 of the 12 group C residues can be observed. (C and D) Details of the three RecUMge residues, i.e., D57, E70, and Q88 (the putative catalytic triad residues), which are potentially involved in coordination of a divalent cation within the catalytic site. Residue L92, located nearby, which is also a group C residue, is clearly visible. (E and F) Overview of the putative catalytic site of RecUMge as viewed from a different angle from that shown in panels A and B. The positions of group C residues D68 and Y66, as opposed to the catalytic triad residues, is illustrated. In panel F, part of the side chain of residue Q88 can just be seen in the slab view of the model. In panel E, this residue cannot be discerned.