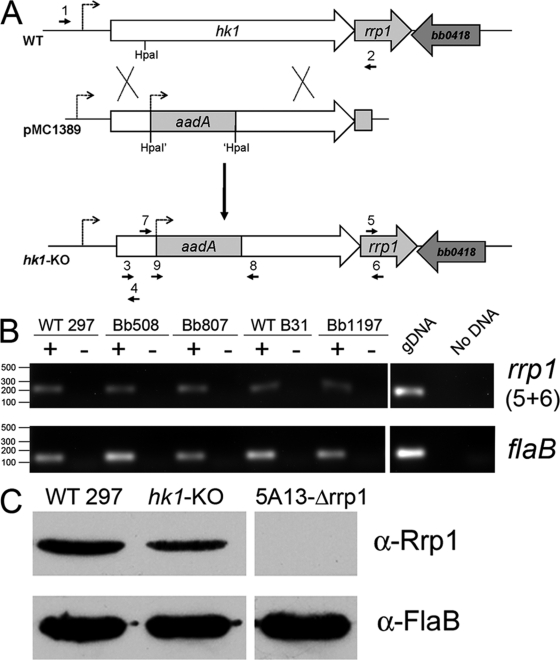
Fig. 2.
(A) Strategy for inactivation of hk1. The hk1 coding sequence plus upstream and downstream flanking regions was amplified from strain 297 using primers ups.hk1-5′ and dwns.hk1-3′ (1 and 2). The hk1 coding sequence was disrupted by insertion of a PflgB-aadA antibiotic resistance cassette into an HpaI restriction site present within the endogenous hk1 gene, yielding pMC1389. Only the relevant portion of pMC1389 is shown. Insertion of the hk1 KO allele was confirmed in strain 297 and B31 mutant isolates using primers hk1-KO-5′ and hk1-KO-3′ (7 and 8) (see Fig. S2 in the supplemental material). Primer sequences are provided in Table 2. (B) Hk1-deficient spirochetes continue to express Rrp1. Semiquantitative RT-PCR was performed on RNAs isolated from a wild-type (WT) (297 and B31) and hk1 KO strains (Bb508, Bb807, and Bb1197) using primers specific for rrp1 (5 and 6) and flaB. RT indicates the absence (−) or presence (+) of reverse transcriptase in the reaction mixture. Purified genomic DNA (gDNA) was used as positive controls. (C) Detection of Rrp1 by immunoblotting. Whole-cell lysates of CE162 (WT 297), Bb508 (hk1-KO), and a previously characterized B31 5A13 Δrrp1 isolate (58) were immunoblotted with rat polyclonal antisera directed against Rrp1 (58) with FlaB used as a loading control. α, anti.