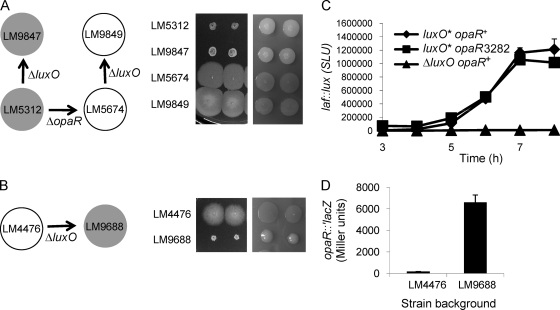
Fig. 4.
Epistasis tests with luxO and opaR alleles. (A and B) (Left) Schemes of strain construction. Arrows indicate strain lineages; filled circles, strains with an OP colony type; open circles, strains with a TR colony type. (Right) Strains were grown on HI media permissive or restrictive for swarming (with low or high agar concentrations) (left and right panels, respectively). Shown are the swarming motilities and colony morphologies of opaR+ and ΔopaR strains (A) and of LM4476 (B) with and without the ΔluxO allele. (C) laf::lux gene expression in the LM1017 (luxO*) background. Strains LM1017 (luxO* opaR+ laf::lux), LM9736 (luxO* opaR3282 laf::lux), and LM9757 (ΔluxO opaR+ laf::lux) were grown on plates and were harvested periodically in order to measure luminescence, reported as SLU. The introduction of an opaR mutation into strain LM1017 to produce LM9736 had no significant effect on luminescence, whereas the luminescence profile of LM9757, which was derived from LM1017 by introducing the ΔluxO allele, was significantly different from those of the other strains (P < 0.001 at all time points). Error bars represent standard deviations of triplicate measurements in one representative experiment of three. (D) The introduction of the ΔluxO allele into LM4476 increases the level of OpaR translation. A translational fusion of lacZ to opaR was introduced into the chromosomes of LM4476 and LM9688, generating strains LM9752 and LM9903, respectively. β-Galactosidase activity was measured over a time course of growth on HI plates and is reported as Miller units at 6 h. The difference between strains was statistically significant (P < 0.0001). Error bars represent standard deviations of triplicate samples in a single representative experiment. The experiment was repeated three times, and the results of a representative experiment are shown.