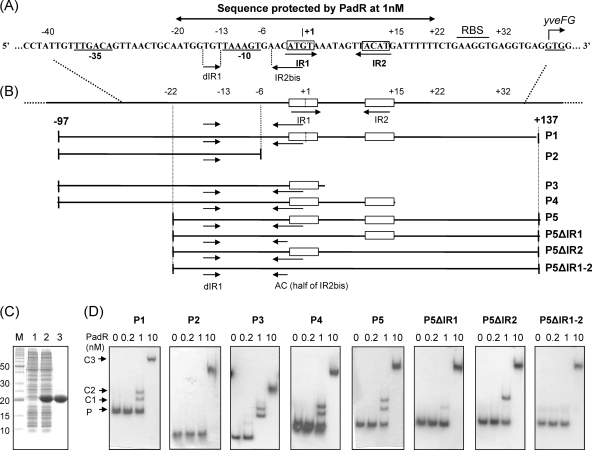
Fig. 3.
EMSA for PadR with native and modified padC promoters. (A) padC promoter DNA sequence indicating nt positions from the +1 transcription start site, −35 and −10 boxes, and IR1-IR2 and dIR1-2bis, the inverted repeat sequences that form the dIR1-2bis/IR1-2 pattern. The start codon (GTG) of yveFG, a nonfunctional gene cotranscribed with padC (32), is indicated. (B) Map of native (P1) and modified (P2 to P5 and P5 derivatives) padC promoters used in EMSA. (C) SDS-PAGE of protein extracts containing purified PadR used in EMSA. Lane M, molecular mass standard; lanes 1 and 2, crude extracts from E. coli BL21 and E. coli BL21/pER, respectively; lane 3, purified PadR. (D) EMSAs with native and modified padC promoters (0.2 nM). PadR was used in EMSAs at concentrations ranging from 0 to 10 nM. A concentration of 1 nM PadR gave specific binding (C1 and C2 complexes), while 10 nM PadR gave unspecific binding (C3 complex). P, unbound probe. For panel D, all PAGE experiments were run under the same conditions, and the fact that the probes and the complexes in the different panels are not aligned horizontally results from the different sizes of the probes (P1, 234 bp; P2, 91 bp; P3, 102 bp; P4, 112 bp; P5, 159 bp; P5ΔIR1 and P5ΔIR2, 155 bp; and P5ΔIR1-2, 151 bp).