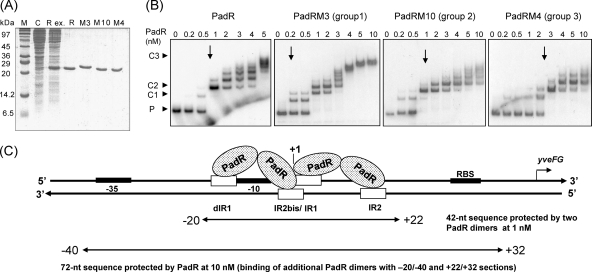
Fig. 5.
(A) SDS-PAGE of native (R) and modified (M3, M10, and M4) His6-tagged PadRs produced in E. coli and used in EMSA. M, molecular mass standard; C, control E. coli strain with pET28a+; Rex, crude extract of native PadR. The same types of extracts were produced to purify the native protein and M3, M10, and M4, shown in the corresponding lanes. (B) EMSAs of native and modified PadRs (M3, M10, and M4), representing the three groups based on PAD activity with the native padC promoter probe at 0.2 nM (see Fig. 4). P, promoter DNA probe without PadR. C1 and C2 indicate specific complexes formed between the DNA probe and PadR. C3 results from additional low-affinity or less specific binding. Arrows indicate the concentrations at which the specific complexes C1 and C2 are observed. (C) Model for PadR dimer binding to padC (yveFG) promoter. The protected sequence was deduced from the results presented in Fig. 2. One dimer binds to dIR1 and the complementary sequence of IR2bis, and the second dimer binds to IR1 and the complementary sequence of IR2.