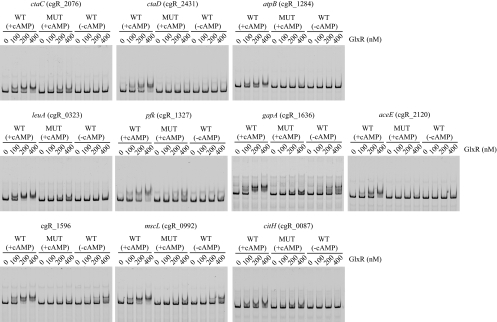
Fig. 2.
EMSAs of representative in vivo GlxR-binding promoter regions detected by ChIP-chip analyses. The regions containing a wild-type (WT) or mutated (MUT) GlxR site were amplified by PCR using primers listed in Table S1 in the supplemental material and were labeled with Cy3 as described in Materials and Methods. The DNA probe was incubated with the purified GlxR at the indicated concentrations in the presence (+cAMP) or absence (−cAMP) of 0.5 mM cAMP and analyzed by nondenaturing PAGE. The target genes are indicated at the tops of the gels. Each well contained 10 nM DNA fragment.