Fig. 1.
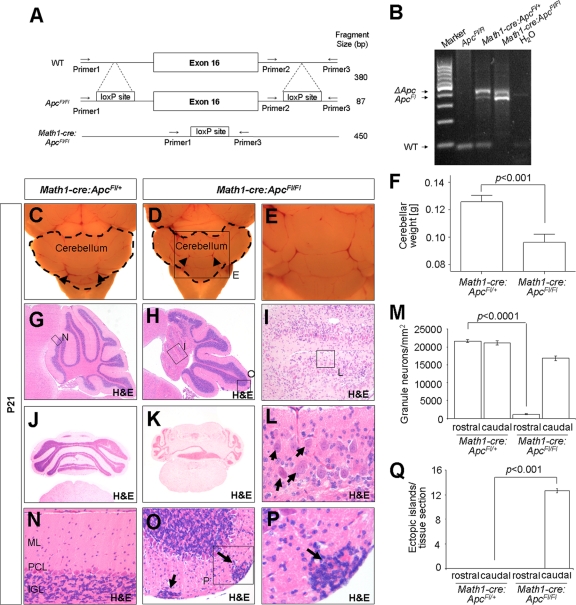
Math1-cre::ApcFl/Fl cerebella displayed severe histomorphological abnormalities. The scheme shows the Apc wild-type (WT) allele, as well as the Apc allele with loxP sites before and after exposition to Cre recombinase (A). PCR of isolated cerebellar DNA from P7 mice was carried out using all three primers indicated in panel A and showed a band for the floxed allele (380 bp) and for the recombined allele (450 bp) in Math1-cre::ApcFl/Fl and Math1-cre::ApcFl/+ mice but not in the wild-type situation (B). In the wild-type situation, primers 2 and 3 amplified an 87-bp fragment, while the DNA fragment between primers 1 and 3 was not amplified due to its large size. Size and weight of P21 Math1-cre::ApcFl/Fl cerebella (n = 6) were largely diminished compared to those for controls (n = 4; P < 0.001, Student's t test). All values were normalized to total brain weight (C to F). Arrowheads in panels C and D delineate corresponding anatomic landmarks. H&E histology revealed a dramatic depletion of granule neurons and a severely disrupted cortical architecture in rostral parts of Math1-cre::ApcFl/Fl cerebella compared to findings for controls both in sagittal (G to M and L to N) and in horizontal sections (J and K). Note the failure of Purkinje cells to form the typical monolayer in cerebella with Apc-deficient granule cells (arrows in panel L). Hypomorphic caudal areas revealed clusters of granule cells that failed to migrate inwardly (O to Q, arrows). IGL, internal granular layer; ML, molecular layer; PCL, Purkinje cell layer; WM, white matter. Enlarged views of boxed areas in panels D, G, H, I, and O are presented in other panels, as indicated.