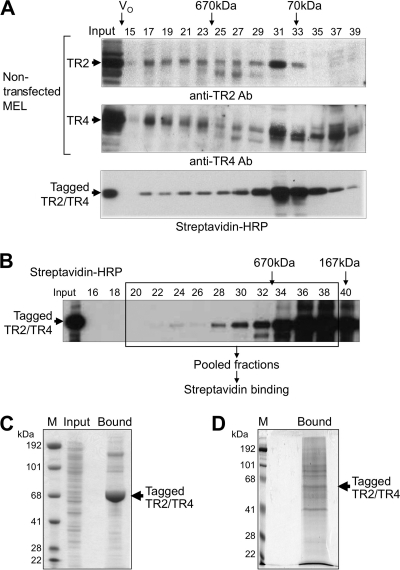
Fig. 3.
Recovery of proteins that copurify with biotinylated TR2/TR4. (A) Size fractionation of nuclear extracts by analytical Superose 6 gel filtration. Fractions from induced (wild-type) MEL cells were analyzed by immunoblotting with anti-TR2 (top panel) or anti-TR4 (middle panel) antibodies. Fractions from induced MEL cells transformed with biotin-tagged TR2 and TR4 were analyzed by affinity blotting with HRP-conjugated streptavidin (bottom panel). The void volume (V0) and the elution of two molecular mass markers are indicated with arrows. (B) Fractionation of biotin-tagged TR2 and TR4 on a preparative Superose 6 column. Biotinylated TR2/TR4 proteins were detected by affinity blotting with HRP-streptavidin. Fractions that were pooled for streptavidin binding and mass spectrometric analysis are boxed. (C) Coomassie blue-stained SDS-polyacrylamide (4 to 12% gradient) gel of proteins bound to streptavidin beads from unfractionated MEL nuclear extract. Unfractionated nuclear extract prepared from induced MEL cells transfected with tagged TR2 and TR4 was incubated with streptavidin beads, from which bound proteins were eluted with 1× Laemmli sample buffer for SDS-PAGE. The arrow indicates the migration position of tagged TR2 and TR4 proteins. Input, MEL nuclear extract before binding to streptavidin beads; M, molecular mass markers. (D) Coomassie blue-stained SDS-polyacrylamide (8%) gel showing proteins bound to streptavidin beads from size-fractionated MEL nuclear extract. Proteins in size-fractionated nuclear extract from the transfected MEL cells were bound to and then eluted from streptavidin beads prior to SDS-PAGE.