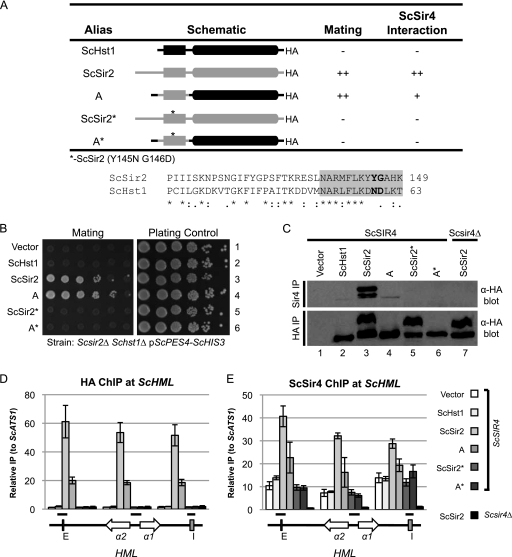
Fig. 7.
Mutations that disrupt the ScSir2-ScSir4 interaction are identified. (A) Summary of mutations and their properties. An alignment of the relevant portion of ScSir2 and ScHst1 is shown. Positions that were mutated in ScSir2 are in bold. The gray box indicates aa 137 to 149, which are essential for ScSir2 function. (B) ScSir2-mediated silencing was assessed using a mating assay. A Scsir2Δ Schst1Δ mutant strain (LRY2083) was transformed with the empty vector or plasmids expressing the constructs in panel A. Mating was assessed as described in the legend to Fig. 2B. (C) The association of the mutant proteins with ScSir4 was examined by co-IP. ScSir4 was immunoprecipitated from the same strains used in panel B or an Scsir2Δ Schst1Δ Scsir4Δ mutant strain (LRY2590) transformed with ScSIR2-HA. (D) The association of ScHst1, ScSir2, and chimeric proteins with HML was examined by chromatin IP (ChIP). HA epitope-tagged proteins were immunoprecipitated from an Scsir2Δ Schst1Δ mutant yeast strain (LRY2507) transformed with the empty vector or plasmids expressing the constructs in panel A or an Scsir2Δ Schst1Δ Scsir4Δ mutant strain (LRY2590) transformed with ScSIR2-HA. (E) The association of ScSir4 with HML in the same strains as in panel D was assessed by ChIP.