Fig. 8.
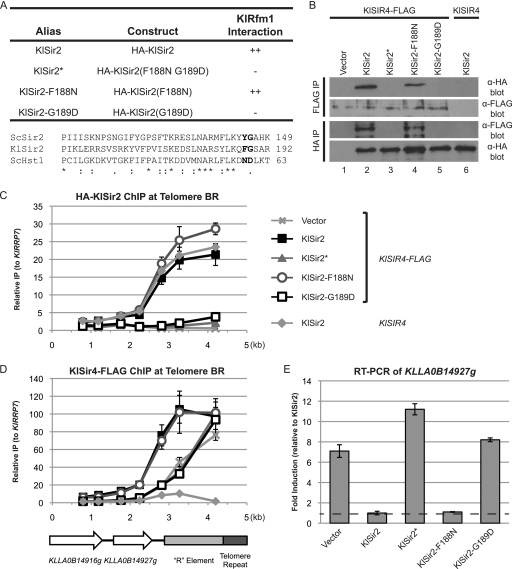
The interaction domain for Sir4 is conserved between KlSir2 and ScSir2. (A) Summary of mutations and their properties. An alignment of the relevant portions of ScSir2, KlSir2, and ScHst1 is shown. Positions mutated in KlSir2 are in bold. (B) The association of HA-KlSir2 with KlSir4-FLAG was examined by co-IP. HA-KlSir2 and KlSir4-FLAG were immunoprecipitated from a Klsir2Δ KlSIR4–FLAG mutant strain (LRY2388) transformed with the empty vector or plasmids expressing the constructs in panel A or from a Klsir2Δ KlSIR4 mutant strain (LRY2333) transformed with HA-KlSIR2. (C) The distribution of HA-KlSir2 at Telomere BR was assessed in the same strains as in panel B by ChIP. (D) The distribution of KlSir4-FLAG at Telomere BR was assessed as described for panel C. (E) Expression of KLLA0B14927g was assessed by quantitative RT-PCR in the same strains as in panel B. Levels of KLLA0B14927g mRNA were first normalized to KlACT1 and then expressed relative to those of the strain containing wild-type HA-KlSir2.