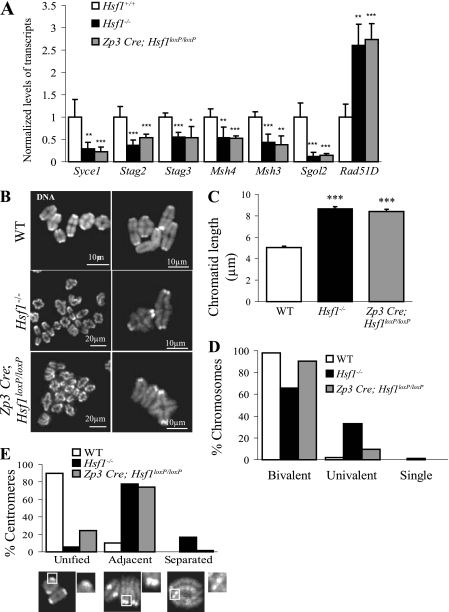
Fig. 7.
Postnatal deletion of Hsf1 (Zp3 Cre; Hsf1loxP/loxP) partially rescues the defects observed in MI chromosomal structure. (A) RT-qPCR performed on adult fully grown oocytes obtained from Zp3 Cre; Hsf1loxP/loxP females revealed that HSF1 target genes are as affected as those in the constitutive knockout. Relative quantities of mRNA were normalized against the quantity of the ribosomal S16 transcripts and their relative expression levels were compared to those of the WT sample, which was arbitrarily given the value 1. Bars represent means ± SEM from at least three independent experiments performed with oocytes collected from several females (n = 3). (B) Representative images of MI chromosome shown at two different magnifications (TO-PRO-3 staining). (C) Chromatid length, measured from images as shown in panel B by ImageJ software, was similarly increased in Hsf1−/− (n = 103) and Zp3 Cre; Hsf1loxP/loxP (n = 108) oocytes compared to WT (n = 112) oocytes. (D) MI chromosomes (as shown in panel B) were classified as bivalent, univalent, or single (number of analyzed chromatids, WT, n = 880; Hsf1−/−, n = 1,494; Zp3 Cre; and Hsf1loxP/loxP, n = 1,032). (E) Proportions of unified, adjacent, and separated sister centromeres in WT, Hsf1−/−, and Zp3 Cre; Hsf1loxP/loxP oocytes (number of centromeres analyzed per genotype, n = 100). *, P < 0.05; **, P < 0.01; ***, P < 0.001.