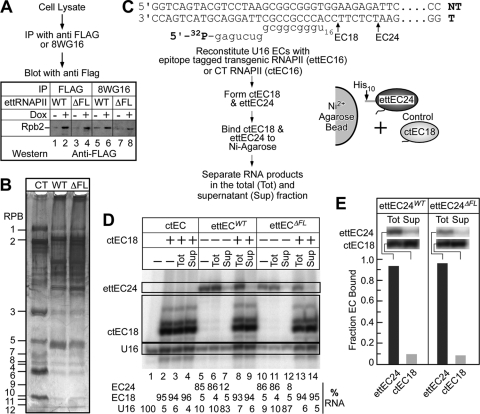
Fig. 2.
Conditional expression, partial purification, and in vitro transcriptional activities of wild-type and ΔFL ettRNAPII. (A) Conditional expression of wild-type and ΔFL ettRNAPII in HEK 293 cells. Cell lysates from cultures induced with doxycycline (2 μg/ml) were immunoprecipitated with either anti-Flag antibody M2 (lanes 1 to 4) or RPB1-specific 8WG16 antibodies (lanes 5 to 8). The immunoprecipitated proteins were fractionated on an SDS–4 to 12% polyacrylamide gel and were detected by immunoblotting. Anti-Flag antibodies were used to detect the expression of the ettRPB2 subunit and its assembly into RNAPII. (B) Partial purification of wild-type and ΔFL ettRNAPII. Flag-tagged wild-type and ΔFL RNAPII were affinity purified on anti-Flag M2 agarose, fractionated on an SDS–4 to 16% polyacrylamide gel, and stained with silver. Purified calf thymus (CT) RNAPII served as a marker. The positions of the 12 subunits of RNAPII are indicated. (C) Sequences of the DNA and RNA oligonucleotides used to reconstitute ECs. The positions of the halted EC18 and EC24 are indicated. Transcription elongation complex reconstitution, EC18 and EC24 formation, and immobilization of ECs on Ni2+ agarose beads are shown schematically. (D) Purified wild-type and ΔFL ettRNAPII are transcriptionally active and are not contaminated with endogenous wild-type RNAPII. Reconstituted wild-type EC16 and ΔFL ettRNAPII EC16 were elongated in the presence of ATP and GTP to make G24 ECs (ettEC24). Control ctEC18 was made using purified CT RNAPII and was mixed with either wild-type or ΔFL ettRNAPII. The complexes were then immobilized on Ni2+ agarose beads and were separated by centrifugation, and the RNA in the total (Tot) and supernatant (Sup) fractions was separated on a 20% denaturing polyacrylamide gel. (E) The fractions of ettEC24 and ctEC18 bound to Ni2+ agarose beads were determined, establishing that >90% of the transcriptional activity in the ettRNAPII preparations was derived from the tagged enzymes and not from native, untagged RNAPII, which could potentially contaminate the preparations.