Fig. 1.
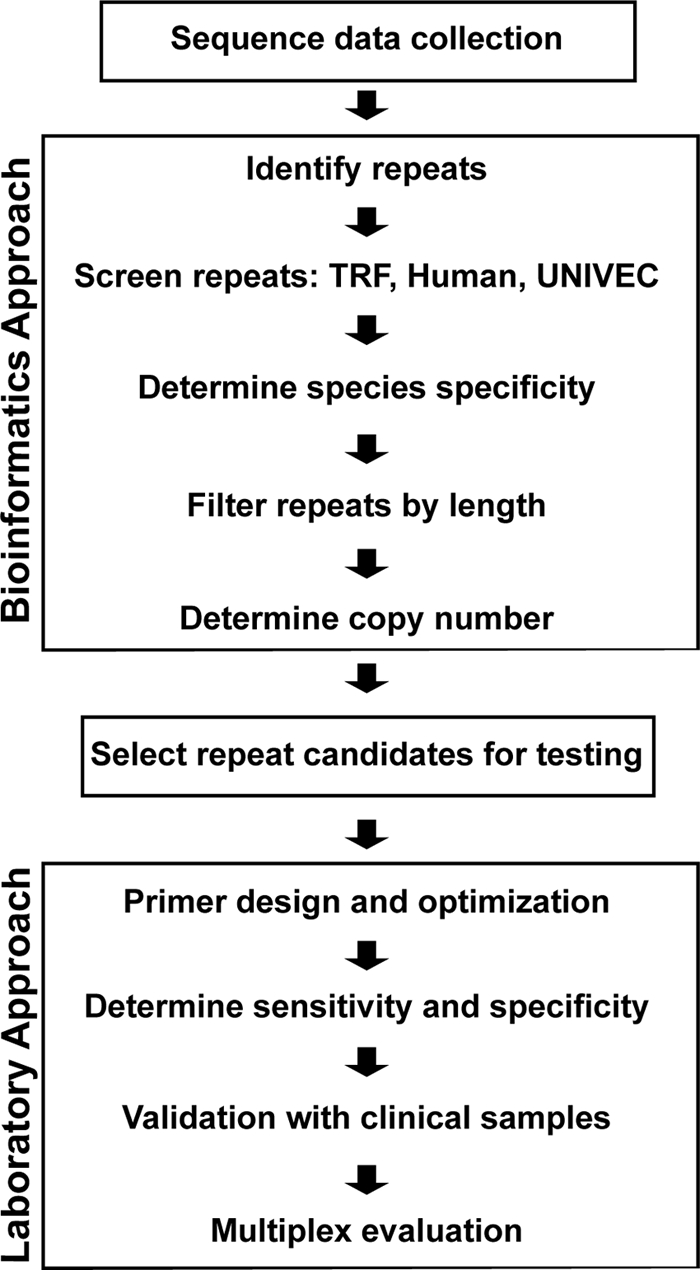
Schematic of diagnostic target screening and development pipeline. All genomic sequences for P. vivax and P. falciparum were downloaded from PlasmoDB. Data were mined for repeats using the RepeatScout algorithm to construct consensus repeat sequences (CRS) for each identified repeat family. CRS were then screened in parallel for tandem repeats, similarity to human sequences, and vector sequences. Any CRS failing these screens were removed from further consideration. CRS that were not species specific or less than 300 bp long were eliminated. Family copy numbers for the remaining candidates were determined via comparison of the CRS against the appropriate genome data. Candidate repeat families containing 6 or more copies separated by at least 100 bp were considered for further testing. For additional information and clinical sample validation, see Materials and Methods.
