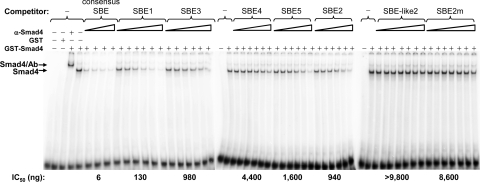
Fig. 5.
Competition EMSAs used to determine relative binding affinities of Zp SBEs for Smad4. Purified GST-Smad4 (100 ng) was incubated with 8 ng of radiolabeled double-stranded consensus SBE and various amounts of the indicated unlabeled double-stranded DNAs as competitors. Immunoshift assays were performed with 2 μg of antibody specific to Smad4. Locations of the protein/DNA complexes are indicated. Amounts of unlabeled double-stranded competitors used were the following: 5, 10, 20, and 40 ng for consensus SBE; 40, 100, 250, 625, 1,562, and 3,906 ng for SBE1, SBE2, SBE3, SBE4, and SBE5; and 40, 100, 250, 625, 1,562, 3,906, and 9,765 ng for SBE-like2 and SBE2m. The IC50s shown below the gels were determined from the concentrations of unlabeled oligonucleotide required to inhibit the binding of GST-Smad4 to the radiolabeled consensus SBE probe by 50%.