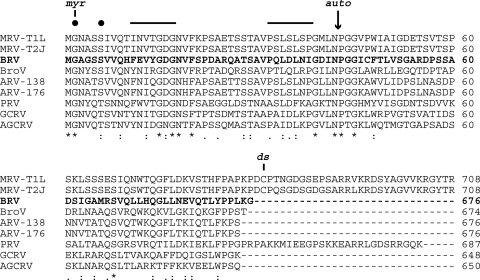
Fig. 3.
Multiple-sequence alignment of ortho- and aquareovirus outer shell proteins including the newly determined sequence from BRV (bold). N-terminal (top) and C-terminal (bottom) portions of the alignment are shown. The alignment was generated using the program Clustal W2 as implemented at http://guidance.tau.ac.il/, using sequences as identified in Materials and Methods. The number at the right end of each line indicates the position of the last amino acid in the sequence. Identities (*), strong similarities (:), and weaker similarities (.) in the aligned sequences are indicated at bottom. Other annotations are for the N-terminal myristoylation site (myr), the conserved myristoylation consensus sequence (filled circles), the putative transmembrane β-strands (solid lines at top) described in the text, the autolytic cleavage site (auto, arrow), and the disulfide-bonding Cys residue of MRVs (ds).