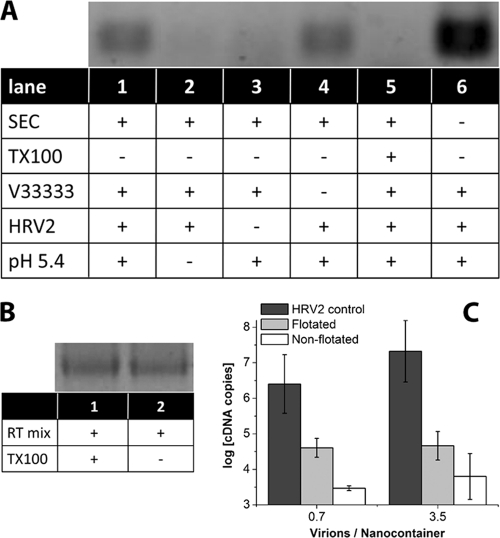
Fig. 3.
Viral RNA is transferred into nanocontainers upon incubation at low pH. (A) HRV2 (0.36 pmol) was bound to 17.4 μl of receptor-decorated nanocontainers (at a virus-to-lipid ratio of 5.6 × 10−6). The mixture was adjusted to 20 μl, brought to pH 5.4 via addition of 1.2 μl 1 M sodium acetate, pH 5.0, and incubated for 15 min at room temperature prior to reneutralization by addition of 0.6 μl 1 M sodium hydroxide. RNA transferred into the liposomal lumen was reverse transcribed by the encapsulated reverse transcriptase at 37°C for 1 h (see scheme in Fig. 1B). Synthesized cDNA was made accessible for subsequent PCR by addition of Triton X-100. The amplified 206 bp fragment was detected by agarose gel electrophoresis. Components present or absent and treatments of the nanocontainers are indicated by (+) and (−), respectively. SEC, not encapsulated RT buffer was removed from the nanocontainers via spin SEC; TX-100, nanocontainers were destroyed with detergent Triton X-100 prior to decoration with receptor; MBP-V33333, the receptor was present on the surfaces of nanocontainers; HRV2, virus was added; pH 5.4, the outside medium was brought to acidic pH. (B) In order to exclude inhibition of the RT-PCR by TX-100, reverse transcription and amplification were carried out by using in vitro-transcribed viral RNA in the presence (lane 1) and in the absence (lane 2) of the detergent. Note that the signal strength is identical. (C) RNA transfer into nanocontainers (without receptor decoration) was triggered as described for panel A, 1 aliquot was directly subjected to qRT-PCR, and 1 was first subjected to flotation. For comparison, the number of RNA copies released from virus into the medium on incubation at pH 5.4 in the absence of nanocontainers is also depicted (HRV2 control).