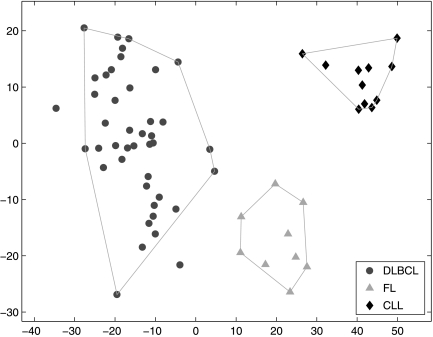
Fig. 1.
SPACC applied to B-cell lymphoma gene expression data, example 1 in Results. The horizontal and vertical axes are the coefficients of the first two principal components. The shape of the nodes represents the input class labels. SPACC divides the samples into three subsets, which are indicated by the polygons. The resulting subsets are consistent with the input class labels.