Figure 1.
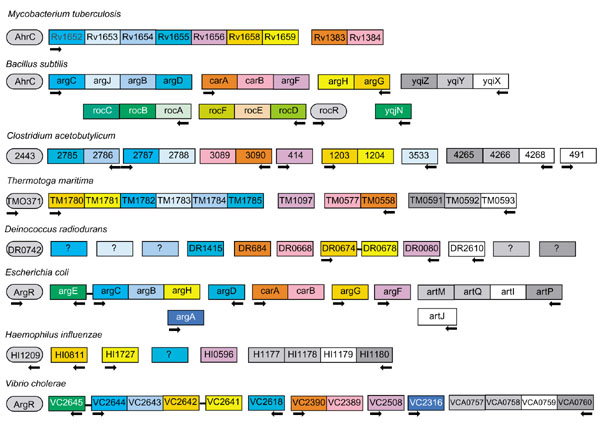
Schematic representation of the operon organization and regulation of the arginine metabolism and transport genes. Genes are represented by boxes. ARG boxes in the upstream region are shown by black arrows. The direction of the arrow indicates the direction of transcription. The linear pathway (in E. coli and V. cholerae) involves N-acetylglutamate synthase (argA) and N2-acetylornithine deacetylase (argE). The circular pathway (in other bacteria) involves N2-acetyl-L-ornithine: L-glutamate acetyltransferase (argJ). The common genes are acetylglutamate kinase (argB); acetylglutamate semialdehyde dehydrogenase (argC); acetylornitine delta-aminotransferase (argD); ornithine carbamoyltransferase (argF, argI); argininosuccinate synthase (argG); argininosuccinate lyase (argH); carbamoyl-phosphate synthase (carAB). The H. influenzae genome contains only argH, argG, argF and possibly argD orthologs. There are difficulties in identifying orthologs for argC, argJ and argB in D. radiodurans because there are several paralogous genes encoding proteins that can possibly perform these functions. The B. subtilisroc operons involved in arginine degradation are also regulated by AhrC, as well as anaerobic arginine catabolism genes arcABCD in B. licheniformis [14] (data not shown). The transporter genes are: periplasmic binding protein (white), permease transmembrane protein (light gray), ATPase component (dark gray).
