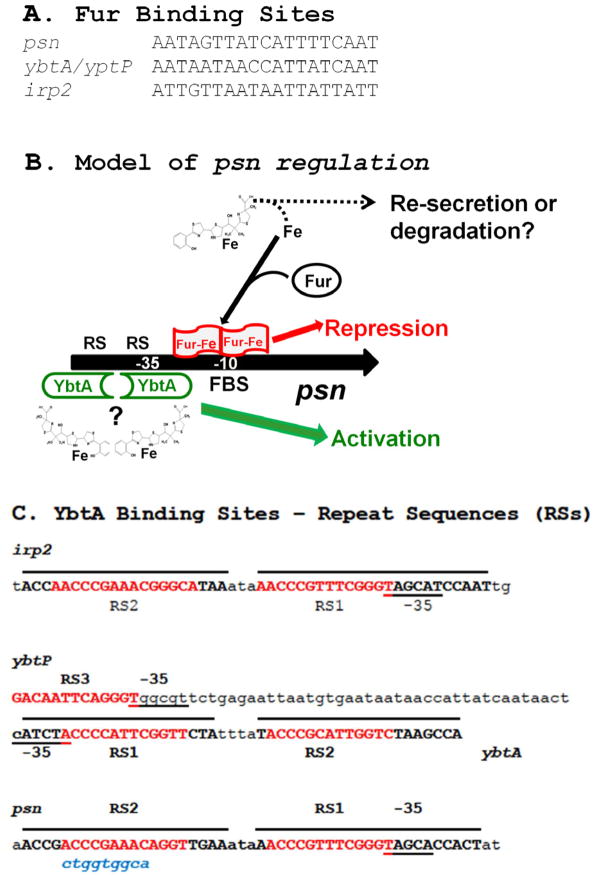
Fig. 5.
Transcriptional regulation by Fur and YbtA. (A) Fur binding sites (FBSs) shown were mapped by Gao et al [50] and the sequences shown are from Y. pestis strain 201. The ybtP FBS is part of the ybtP/ybtA intergenic region and identical to the ybtA FBS. (B) Model of psn regulation. During iron-sufficient conditions, the psn promoter is repressed through Fur-Fe binding to an FBS that overlaps the −10 region. Under iron-deficient conditions this repression is relieved due to a shift to primarily iron-free Fur. If Ybt siderophore is present, YbtA activates transcription of this promoter by binding to repeat sequences (RSs), one of which overlaps the −35 region. The irp2 and ybtP promoters are regulated in the same manner. In contrast, YbtA represses transcription of its own promoter (not shown). Question marks indicate unresolved questions in our model. Our regulatory model favors a mechanism in which Fe-Ybt binds to YbtA for regulatory activity. However, binding of Fe-Ybt or iron-free Ybt to YbtA has not been demonstrated. The model is reproduced with modifications from Miller et al [31] with the permission of the Society for General Microbiology. (C) YbtA binding sites in the irp2, ybtP, ybtA, and psn promoter regions. Repeat sequences (RSs) are involved in YbtA binding. RS1, RS2, and RS3 as defined by Animisov et al are denoted in red, uppercase text [57–58]. Extended RS (overlined uppercase text) are proposed based on comparison of RSs among the ybt promoters. Underlined text indicates −35 regions of the promoter regions. The lowercase blue text below the psn sequence shows the mutated sequence that lowered expression of a psn::lacZ reporter [53].