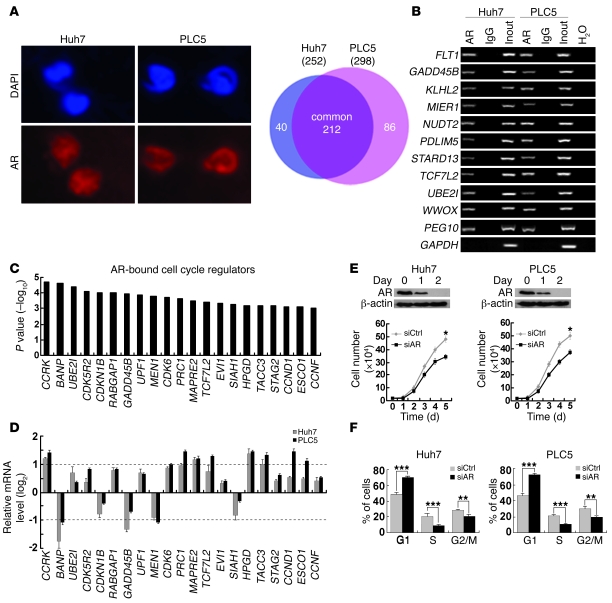
Figure 1. Genome-wide location analysis of AR-binding sites identifies cell cycle–related target genes in HCC cells.
(A) Identification of AR direct target genes using ChIP-chip. In Huh7 and PLC5 HCC cells, AR expression was localized in the nuclei, which were counterstained by DAPI. Venn diagram showing the significant overlap of AR target genes between the 2 cell lines. Original magnification, ×400. (B) Confirmation of 10 randomly selected AR target genes by ChIP-PCR using anti-AR antibody or irrelevant antibody against IgG (negative control) in Huh7 and PLC5 cells. Input (2%) represents the genomic DNA. PEG10 and GAPDH were included as positive and negative controls, respectively. (C) Enrichment of cell cycle regulators in AR target genes as denoted by their highly significant binding P values. (D) Quantitative RT-PCR analysis of the AR-bound cell cycle regulator expressions in Huh7 and PLC5 cells treated with the AR agonist R1881 (100 nM) for 6 hours relative to the untreated cells. GAPDH was used as an internal control. (E) Silencing AR expression retarded HCC cell growth. Western blot analysis of AR following RNA interference. β-actin was used as a loading control. Cell growth was inhibited in Huh7 and PLC5 cells treated with siAR compared with siCtrl-treated cells. (F) G1/S cell cycle progression was inhibited after knockdown of AR expression in both HCC cell lines. *P < 0.05; **P < 0.01; ***P < 0.001. Data are presented as mean + SD of 3 independent experiments.