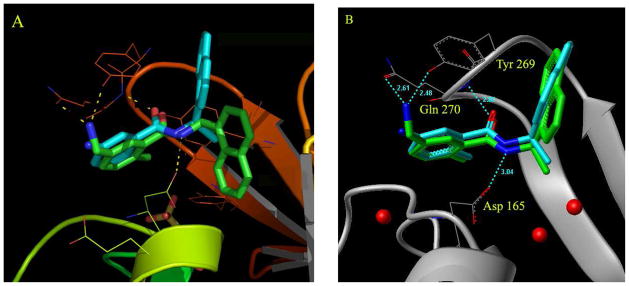
Figure 2.
Docking in the presence of conserved water molecules is critical for replicating the binding conformation of inhibitors in the active site of the bound form of SARS-CoV PLpro. (A) Docking of compound 2 in the absence of water molecules in the active site of the inhibitor-bound form of SARS-CoV PLpro causes the naphthyl rings to flip down into a pocket, as shown by the docked conformation of compound 2 (in green) when compared to the crystal structure conformation of compound 24 (in cyan). In the x-ray structure of compound 24, the naphthyl rings are flipped up in the opposite direction, holding the flexible loop in place. The yellow dotted lines show the possible interactions of the docked compound 2 with residues Tyr269, Gln270 and Asp165 (catalytic domain residue numbering). (B) The three conserved water molecules are marked; two of them are buried deep in the pocket (P5) whereas the third one lies in a groove between residues Lys158 and Glu168. The position of these water molecules is integral to structure-based inhibitor design efforts. The crystal structure conformation of compound 24 is shown in cyan whereas the docked conformation of compound 2 in the presence of water molecules (red dots) is shown in green.