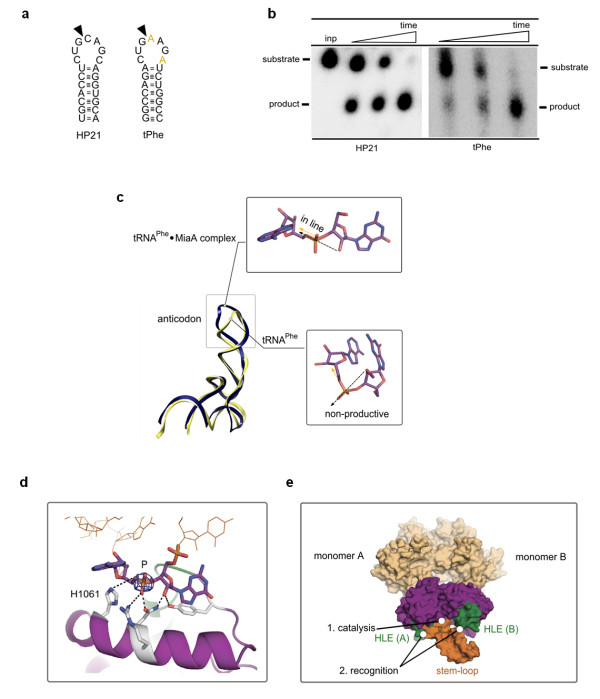
Figure 5.
Model of stem-loop recognition via a composite protein surface. (a) Stem-loops derived from XBP1 mRNA (HP21) and phenylalanine tRNAPhe (tPhe). Loop sequence differences are colored yellow in tPhe. (b) Time courses (0-5 min) for cleavage of 5'-32P-labeled stem-loops HP21 and tPhe by Ire1KR32 (3 μM) analyzed by polyacrylamide gel electrophoresis. (c) Superposition of crystal structures of tRNAPhe (PDB ID 1ehz) and tRNAPhe from tRNAPhe·MiaA complex (PDB ID 2zm5). Black arrow shows direction of nucleophile attack, orange arrow shows direction of leaving group. (d) Manual rigid-body docking of the base-flipped tRNAPhe anticodon into Ire1 crystal structure. Electron density for the scissile phosphate (contoured at 12σ) and the proposed catalytic mechanism (Figure 3c-I) were used as docking constraints. P designates scissile phosphate. The helix-loop element (HLE) is colored green. (e) Surface rendering of the model in (d). The model suggests cleavage of a stem-loop by one active site and recognition by a composite RNA binding surface.