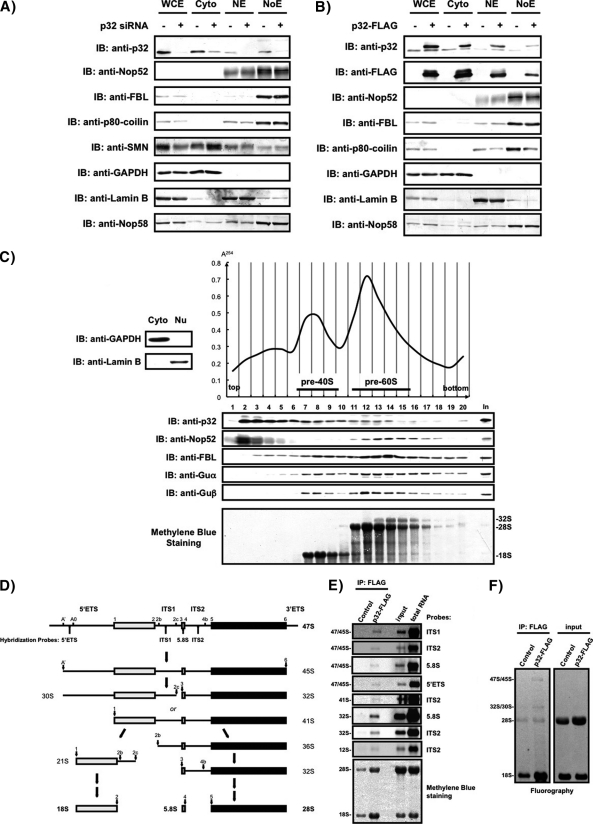
Fig. 4.
The presence of p32 in the nucleolus/Cajal body fraction prepared by cell fractionation and in the preribosome fractions separated by ultracentrifugation. Equivalent amounts of whole-cell extract (WCE), cytoplasmic fraction (Cyto), nucleoplasmic extract (NE), and nucleolar extract (NoE) prepared from 293EBNA cells transfected with (+) or without (–) a p32-specific siRNA (A), or transfected with (+) or without (–) p32-FLAG expression vector (B), were analyzed by immunoblotting with the antibodies indicated at the left. C, Cells were fractionated into nuclear (Nu) and cytoplasmic (Cyto) fractions and were analyzed by immunoblotting (IB) with the antibodies indicated (left, top). The Nu fraction that corresponded to the nuclear extract combining NE and NoE in (A) was separated on 10–30% sucrose density gradients and separated into 20 fractions. The absorbance at 254 nm was monitored and is shown on the top right. Proteins in each fraction were analyzed by immunoblotting with the antibodies indicated at the left (middle). RNAs were also extracted from each fraction, separated by agarose gel electrophoresis, transferred to a membrane, and stained with methylene blue (bottom). Input: 20 μg of protein from unfractionated Nu was loaded for immunoblotting; 2 μg of RNA from unfractionated Nu was loaded for agarose gel electrophoresis and methylene blue staining. D, Structure of the primary rRNA transcript (47S), outline of pre-rRNA processing in mammalian cells adapted from refs. 41, 52, 75, and oligonucleotide probes used for hybridization analysis are indicated. The main processing sites termed 1 to 6 correspond to the ends of mature 18S, 5.8S, and 28S rRNAs are also indicated. The external and internal transcribed spacers (ETS and ITS, respectively) contain additional cleavage sites designated A', A0, 2b, 2c, and 4b. E, RNAs extracted from p32-associated complex were detected by Northern blot analysis with the oligonucleotide probes indicated. Positions of pre-rRNAs and the probes used are indicated on the left and right, respectively. Input: 2 μg of RNA extracted from soluble cell lysates from control cells were loaded. Total RNA: 2 μg of total RNA were loaded. F, RNAs containing newly synthesized pre-rRNAs labeled with [3H]uridine were isolated from p32-associated complex, separated on an agarose gel, transferred to a membrane, and detected by fluorography. Positions of pre-rRNAs are indicated on the left. Input: RNAs (5000 cpm/lane) extracted from soluble cell lysates were loaded.