Fig. 2.
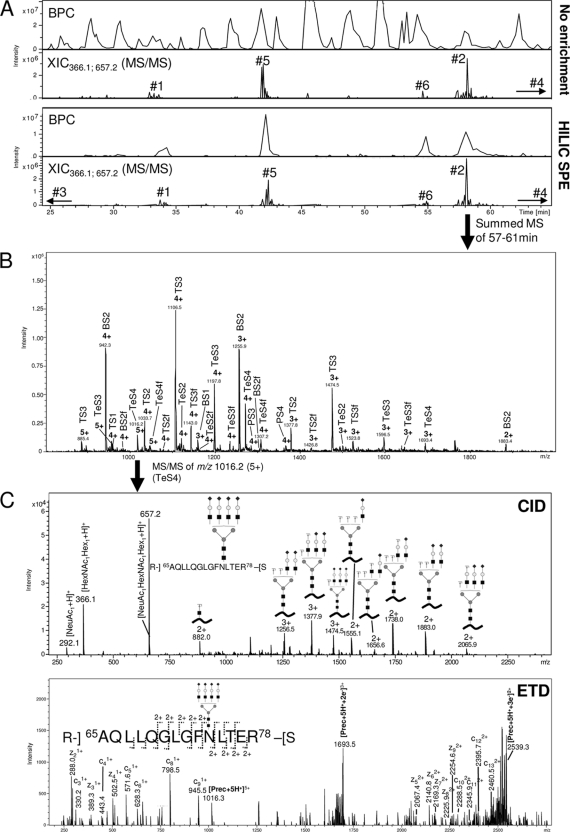
Glycopeptide profiling using LC-MS/MS based glycoproteomics. A, Base peak chromatograms (BPCs) and extracted ion chromatograms (XICs) (m/z 366.1 and m/z 657.2, fragment ions) for LC-MS/MS of non-enriched (top) and HILIC solid phase extraction (SPE) enriched (bottom) peptide mixture generated from a sequential trypsin/Asp-N digest of CBG. Glycopeptide eluting regions and their glycosylation site origins have been marked. A non-biased and close-to-complete isolation of glycopeptides was observed using off-line HILIC enrichment. B, Summed MS full scans of N-glycopeptides originating from glycosylation site 2. A relatively large elution window (i.e. 57–61 min) was allowed to ensure that the complete set of glycoforms was included. See ‘Experimental procedures’ for list of all elution windows. A heterogeneous set of glycoforms was observed (see Table I and Fig. 3 for nomenclature/structures). C, Examples of CID (upper) and ETD (lower) fragmentation of an N-glycopeptide originating from glycosylation site 2 (i.e. Ala65-Arg78 conjugated with a tetra-antennary, fully sialylated complex N-glycan, m/z 1016.2, 5+).