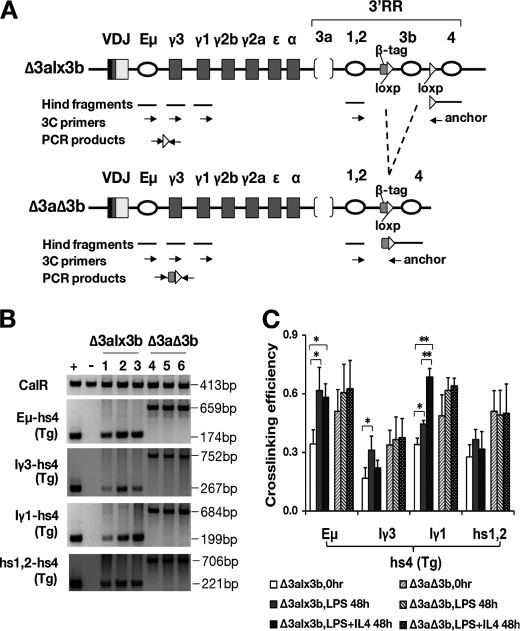
FIGURE 5.
Long range interactions between the 3′RR, IgH transcription unit, and CH region promoters. A, maps of BAC transgenes before (Δ3alx3b) and after (Δ3aΔ3b) hs3b deletion. 5′ primers for Eμ, the Iγ3 promoter, Iγ1 promoter, and 3′RR element hs1.2 are indicated as arrows, as is the transgene-specific 3′ primer near hs4 (anchor). VDJ, inserted VH region. 3C, chromatin conformation capture. B, PCR products generated by chromatin conformation capture (see “Experimental Procedures”) with the indicated primer pairs (left of gel). Interactions between fragments within the CalR gene served as a positive control (top panel). + = positive control template (mixture of CalR fragments and the IgHΔ3alx3b BAC); − = negative control template (mixture of CalR fragments and a BAC with sequences equivalent to the wild-type, endogenous Igh loci). Cell source of template is shown above the lanes (Δ3alx3b and Δ3aΔ3b B cells). Lanes 1 and 4 are from B cells at time 0 (resting B cells); lanes 2 and 5 are from B cells 48 h after LPS stimulation, and lanes 3 and 6 are from B cells 48 h after LPS+IL-4 stimulation. C, summary of cross-linking efficiency data. Cross-linking was measured between the hs4 region of the BAC transgene (hs4(Tg) and the indicated regions upstream (see “Experimental Procedures” for details). Two independent CSR experiments were performed using two different paired mouse lines, and PCRs were performed at least twice with each template. The first three bars in each set of six (for each primer pair) are the results with IgHΔ3alx3b B cells; the second three bars are with IgHΔ3aΔ3b B cells (see legend). * = p < 0.05; ** = p < 0.01.