FIGURE 8.
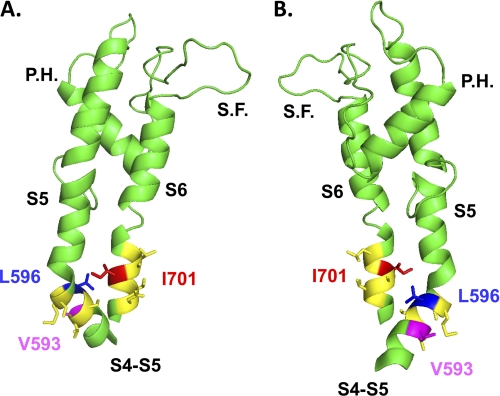
Two different views (A, B) of the computer-based molecular model of domain II of CaV2.3 are shown in a schematic representation. Modeling of domain II was based upon the atomic coordinates of KV1.2 (Protein Data Bank code 2A79) and was achieved with Modeler 9v4 from the S4 to the end of S6. For sake of clarity, only the region going from the S4S5 linker to the end of S6 is shown. The regions in yellow correspond to the regions studied in our double mutant cycle analysis, namely residues Val-593 to Ser-598 in IIS4S5 and Leu-699 to Asp-704 in IIS6. Val-593 and Leu-596 (IIS4S5) are shown in stick representation in magenta and in blue, respectively, whereas Ile-701 (IIS6) is shown in red. The shortest distance between Leu-596 and Ile-701 was estimated to be 9.4 Å from the α-carbon of Leu-596 to the β-carbon of Ile-701. S.F. stands for selectivity filter and P.H. stands for pore helix. The figure was produced using PyMol (DeLano Scientific).