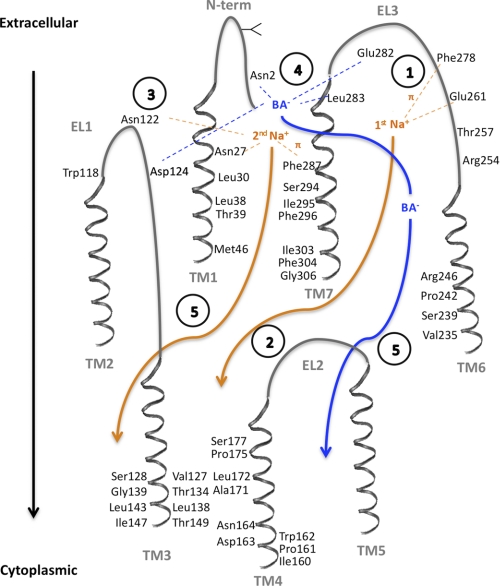
FIGURE 10.
Putative ASBT transport cycle. The proposed mechanism was based on the established 7 transmembrane domain topology (11) and putative 2:1 sodium:substrate stoichiometry (25), as well as current and previous publications. Direct interaction of sodium (orange) or bile acid (blue) with amino acid residues is denoted as dotted lines, while translocation through reputed substrate pathways is shown as full arrows. Transmembrane domains (TM1–7) and extracellular loops (EL1–3) are shown in gray, and amino acid residues involved in substrate interactions, or that line the substrate pathway, are visualized. Intracellular loops are omitted for clarity. Circled numbers describe each proposed step: (1) Stabilization of the 1st Na+ ion within EL3 via the negatively charged sodium sensor Glu261 (16, 19, 20) and cation-π forces with the Phe278 aromatic ring (16); (2) Na+ binding triggers conformational changes that will facilitate its own permeation, and (3) simultaneously favor binding of the sodium sensor Asn122 (19, 20) to the 2nd Na+ ion, which is further stabilized by amino-aromatic/cation- π interactions with Phe287 and the amide proton in Asn27. (4) Structural changes induced by binding/stabilization of the 2nd Na+ will render the binding pocket formed by Glu282, Leu283, and Asn2 (11), and possibly Asp124, available to interact with the bile acid (BA−), leading to coupled Na+ and BA− translocation through translocation pathways in TM1 (current work), TM6 (12) and TM7 (15), and exit into the cytoplasm through TM3 (14, 17) and TM4 (18) (5). Involvement of TM2 and TM5 in substrate/ion binding or translocation remains to be investigated.