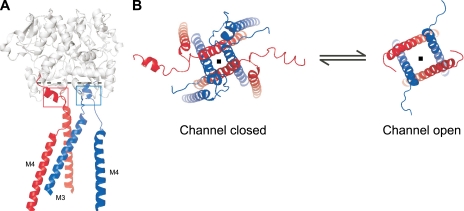
Figure 1.
The M3 transmembrane segment is the major channel-gating element in iGluRs. (A) Backbone structure of two glutamate receptor subunits (GluA2cryst; subunits B and C, Protein Data Bank accession no. 3KG2), each harboring an extracellular LBD (in gray) comprised of polypeptide segments S1 and S2, transmembrane segments M3 and M4, and their associated linkers M3–S2 and S2–M4. For clarity, the M1 transmembrane segment and the M2 pore loop are not shown. GluN1 is assumed to adopt the A/C conformation (red) and GluN2 the B/D conformation (blue) (Sobolevsky et al., 2009). The dashed line indicates point of view (looking down the ion channel) shown in B. Red and blue squares depict regions of intrasubunit cross-linking of the M3–S2 and S2–M4 linkers in GluN1 and GluN2A, respectively. Although there are limitations in comparing NMDA receptors to the AMPA receptor structure and the derived open-state structural model, for example, domain arrangements may be different (Stroebel et al., 2011), we use this information to illustrate general features of gating in iGluRs, specifically for the TMD. (B) Presumed gating movements of the M3 transmembrane segment leading to pore opening. (Left) Tetrameric arrangement of M3/M3–S2 and M4/S2–M4 adopting the A/C (~GluN1; red) and B/D (~GluN2A; blue) conformations in an antagonist-bound channel closed state (Sobolevsky et al., 2009). The M3 transmembrane helices line the channel pore (depicted with a dot), whereas the external helices surrounding this core are the M4 segments. (Right) Reorientation of the M3 segments in the channel open state as predicted from superposition of the GluA2cryst on the closed KcsA and the open Shaker K+ channels (Sobolevsky et al., 2009). In the present study, we restrict these gating rearrangements of M3 through intrasubunit cross-linking of the M3–S2 and S2–M4 linkers in GluN1 (A, red box) or GluN2A (A, blue box).