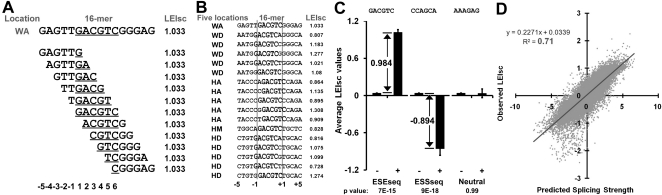
Figure 4.
Contexts created by overlapping 6-mers. (A) An example of how a substitution (GACGTC) creates 11 overlapping WA 6-mers spanning a 16-nt region. The 11 variable 6-mers are distinct for each location and all are assumed capable of influencing splicing. To take the overlapping 6-mers into account, each was assigned the value 1.033, representing the LEIsc value observed for this pre-mRNA molecule. Library bases are underlined. (B) An example of one 6-mer (GACGTC) and the LEIsc values of all the molecules from all locations that contain it in the 16-nt overlap region. (C) Assignment examples for an ESEseq (GACGTC), an ESSseq (CCAGCA), and a neutral 6-mer (AAAGAG). The 20,480 total molecules were classified into two categories: those in which a 6-mer was absent (−) or present (+). Average LEIsc values for each category are shown. Splicing enhancers (ESEseqs) are defined as 6-mers, for which the average LEIsc value is significantly higher when present and its ESEseq score is the difference between the average LEIsc values of the two categories. Splicing silencers (ESSseqs) are defined as 6-mers, for which the average LEIsc value is significantly lower when present. Neutral 6-mers are defined as 6-mers, for which the two values are not significantly different. P-values are from a t-test. The error bar is SEM. (D) Comparison of the observed LEIsc value of a library pre-mRNA molecule with the splicing strength predicted from the additive model described in the text. The chart contains 20,480 points.