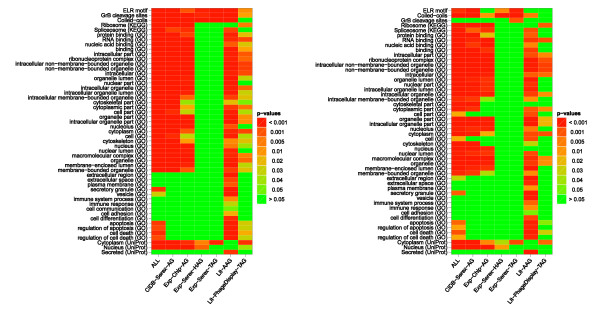
Figure 2.
Heat maps showing the significantly enriched motifs, pathways, GO terms and subcellular locations in the considered data sets. Heat maps showing the significantly enriched motifs, pathways, GO terms and subcellular locations in the considered data sets compared to the ProteinCodingGenes reference set (left-hand side) or compared to the ProteinCodingGenesLongerExons reference set (right-hand side). ALL: union of all antigen sets; CIDB-Serex-AG: retrieved from the Cancer Immunome Database (SEREX method); Exp-Chip-AG: mixed antigens (tumor/non-tumor diseases) identified by protein macroarray; Exp-Serex-HAG: natural occurring autoantigens (SEREX method); Exp-Serex-TAG: tumor antigens (SEREX method); Lit-AAG: autoimmune antigens collected by literature search; Lit-PhageDisplay-TAG: tumor antigens identified by Phage Display experiments collected by literature search