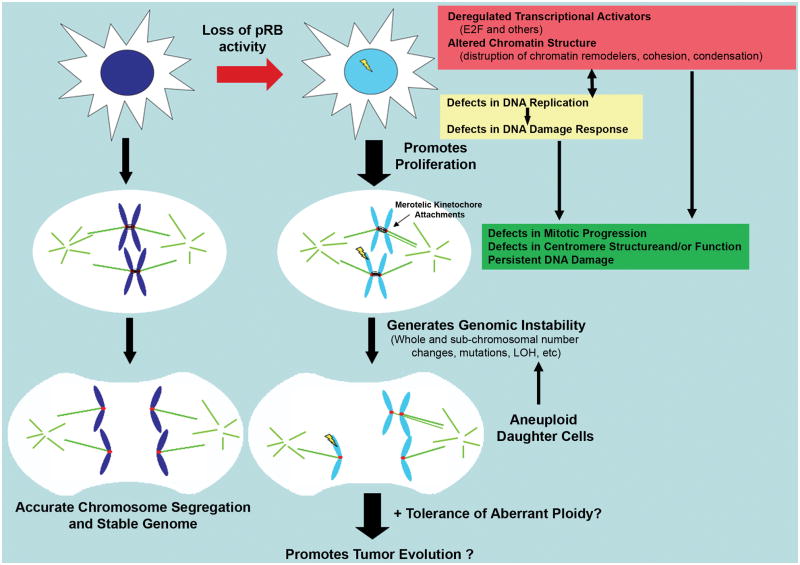
Figure 1. Complex role of pRB in maintenance of genomic stability.
Functional inactivation of pRB has been shown to result in numerous changes, including transcriptional changes (local effects: regulation of specific transcriptional activators: E2F, etc; broad effects: regulation of heterochromatin formation, including telomeric and centromeric regions; and global effects: chromosome condensation and cohesion levels [purple box, top right]). Each of these defects in turn influence DNA replication, mitotic fidelity and the DNA damage response machinery through the transcriptional regulation of important players (yellow and green Boxes; DNA damage represented by yellow lightning bolt). In addition, transcription-independent interactions of pRB with components of many of these pathways have been identified, raising the possibility that pRB’s influence on these downstream processes may be independent of its role in transcription (yellow and green Boxes). Importantly, defects in each of these cellular processes have been shown to result in genomic instability. In addition, pRB loss has been implicated in tolerance of aneuploidy, thereby allowing the propagation of these newly aneuploid cells. The molecular mechanisms behind many of these effects remain unclear and the complex interplay between different effectors of pRB loss makes it difficult to tease apart functional relevance. The importance of individual effects of pRB inactivation under different cellular contexts may in part explain the varying degree of genomic instability seen in tumors possessing pRB pathway lesions.