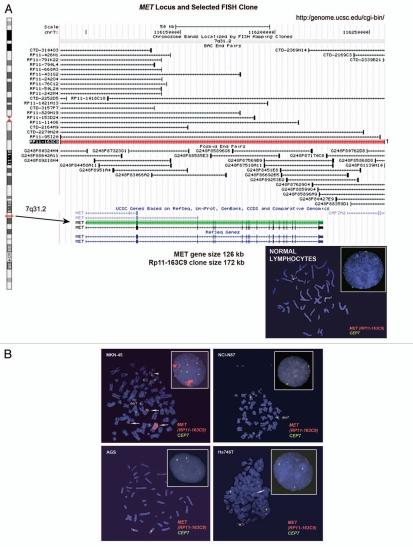
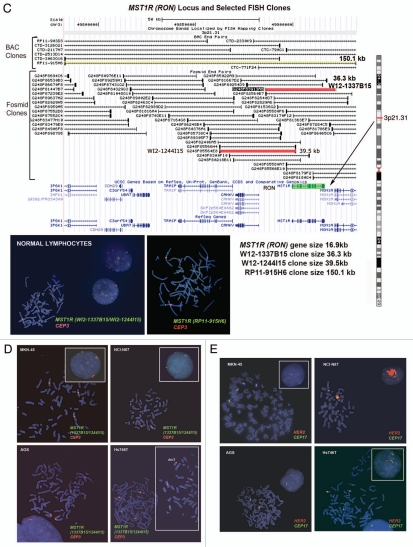
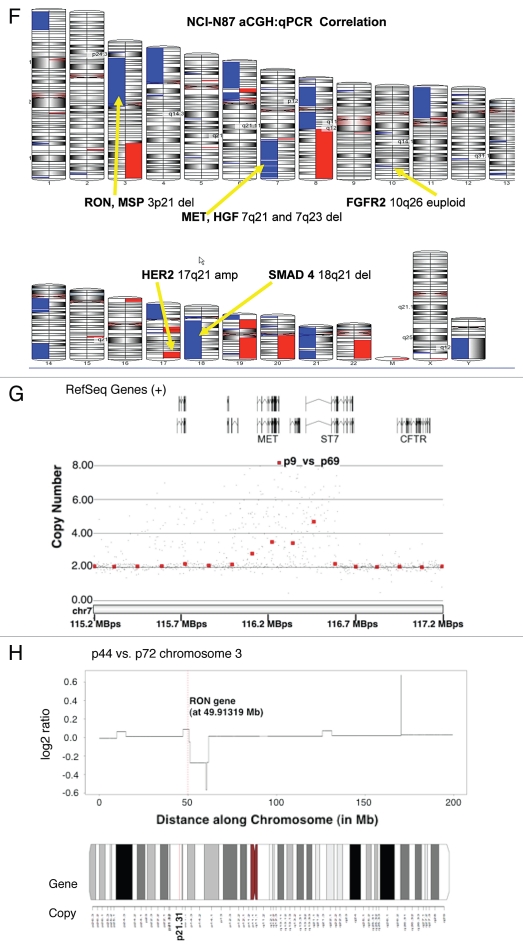
Figure 6.
Alterations in gene copy number of MET, MST1R and HER2 in MKN-45, NCI-N87, AGS and Hs746t GEC cell lines detected by FISH (A–E) and aCGH (F–H). (A) Genomic position of BAC RP11-163C9 clone selected for homebrewed MET FISH probe (top) and representative photomicrograph of MET:CEP7 FISH in control cells from normal human lymphocytes (bottom) in metaphase and interphase (insert) are shown. Here and in (B), the MET gene is localized by red fluorescent signals and chromosome 7 centromere (CEP7) is localized by green fluorescent signals. The cells were counterstained with DAPI (blue). Two signals for each probe can be detected on chromosome 7 as a normal pattern. (B) Images of MET:CEP7 FISH in GEC cell lines in metaphase and interphase (inserts) are presented. MKN-45 cells show multiple red signals representing highly amplified and translocated MET copies on three nonhomologous chromosomes (arrow). Five copies of derivative chromosome 7 (der7) with one der7 carrying MET gene are visible. One copy of chromosome 7 with presumably normal MET signal was also detected (arrowhead). In NCI-N87 cells (top right part), one copy of MET and two copies of CEP7 are visible. MET was deleted in 87% of cells. FISH image of AGS cell line (bottom left) depicts two copies of presumably normal MET and CEP7 signals that were detected in 90% of cells. Three copies of chromosome 7 and four copies of MET (one copy translocated to nonhomologous chromosome) represent Hs746t cells (bottom right). (C) Genomic position of BAC RP11-915H6 clone and two fosmid clones WI2-1337B15 and WI2-1244I15 selected for homebrewed MST1R FISH probe are shown (top). Images of MST1R:CEP3 FISH in control normal human lymphocytes in metaphase and interphase (insert) are presented (bottom). The MST1R gene is detected by green fluorescent signals and chromosome 3 centromere (CEP3) is detected by red fluorescent signals. The combination of fosmid clones (bottom left) gave optimal probe size that was specific to MST1R DNA, and gave bright FISH signals comparable to BAC probe signals (bottom right). Normal pattern of two signals for each, MST1R and CEP3 probes can be detected on chromosome 3. (D) Images of MST1R:CEP7 FISH in GEC cell lines in metaphase and interphase (inserts) are presented. MKN-45 cells show three copies of each, MST1R and CEP3 (balanced trisomy; top left). One copy of MST1R and two copies of CEP3 are visible in NCI-N87 cells (top right part). MST1R was deleted in 83% of these cells. AGS cell line carries two copies of each presumably normal MST1R and CEP3 signals (bottom left). Hs746t cells revealed heterogeneous pattern of alterations of MST1R and chromosome 3 signals (bottom right). Image of Hs476t shows cell with balanced tetrasomy for both signals and partial metaphase spread in insert with three MST1R and four CEP3 signals. (E) Images of HER2:CEP17 FISH in GEC cell lines in metaphase and interphase (including inserts in top left and bottom right) are presented. The HER2 gene is localized by red fluorescent signals and chromosome 17 centromere CEP17 is localized by green fluorescent signals. MKN-45 HER2-nonamplified cells show balanced tetrasomy for both signals (top left). NCI-N87 show classical example of true gene amplification forming intact amplicon on one chromosome 17; another, presumably normal chromosome 17, carries one copy of HER2 (top right). On the bottom left, FISH image of AGS cell line depicts two copies of presumably normal HER2 and CEP17 signals. Hs746t cells were HER2 FISH− and show three copies of each HER2 and chromosome 17 (balanced trisomy; bottom right). Correlation of qPCR and aCGH (Agilent 1M array normalized to pooled normal lymphocyte DNA) gene copy number results for NCI-N87. Gene copy number by qPCR are indicated (F). aCGH copy number (blue, deletion; red, gain) See Table 1 in text for details. (G) Evaluation of genome copy number by aCGH; chromosome 7 results are shown from tumor sample (p9) competitively hybridized with adjacent grossly normal tissue sample (p69) revealing increased MET copy number in the tumor. (H) Evaluation of genome copy number by aCGH; chromosome 3 results are shown of tumor sample (p44) competitively hybridized with adjacent grossly normal tissue sample (p72) revealing increased RON copy number (trisomy) in the tumor, adjacent to a large deleted region.