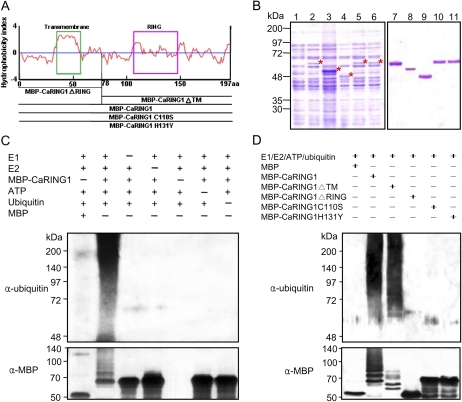
Figure 3.
E3 ubiquitin ligase activity of CaRING1. A, Hydrophobicity index and expression constructs used for the in vitro ubiquitination assay. Recombinant fusion proteins are as follows: MBP-CaRING1, containing the full-length CaRING1; MBP-CaRING1ΔRING and MBP-CaRING1ΔTM mutants, with deletion of the C-terminal RING-H2 domain and N-terminal TM domain, respectively; and MBP-CaRING1C110S and MBP-CaRING1H131Y substitution mutants, with replacement of Cys-110 with Ser and His-131 with Tyr, respectively. The green box indicates the TM domain. B, SDS-PAGE of the recombinant MBP-CaRING1 and mutant variant proteins. Fusion constructs were transformed into E. coli BL21 (DE3). Cells were grown in LB medium, and expression of recombinant proteins was induced with 300 μm isopropyl-β-d-thiogalactopyranoside. Lane 1, Soluble fraction of uninduced E. coli MBP-CaRING1 extract. Lanes 2 to 6, Soluble fractions of induced E. coli MBP-CaRING1, CaRING1ΔTM, CaRING1ΔRING, CaRING1C110S, and CaRING1H131Y, respectively. Induced proteins are indicated by asterisks. Lanes 7 to 11, Purified MBP-CaRING1, CaRING1ΔTM, CaRING1ΔRING, CaRING1C110S, and CaRING1H131Y, respectively. C, E3 ubiquitin ligase activity assay of CaRING1. Recombinant MBP-CaRING1 fusion protein was incubated in the presence or absence of E1 (ScUBA1), E2 (AtUBC10), ATP, and/or ubiquitin. The reactions were analyzed with immunoblots using anti-ubiquitin antibodies (top panel) and anti-MBP antibodies (bottom panel). E3 ubiquitin ligase activity of MBP-CaRING1 was only detected in the presence of E1, E2, ATP, and ubiquitin. D, E3 ubiquitin ligase activity assay of CaRING1 mutant proteins. MBP-CaRING1 protein was used as a positive control. Only the mutant MBP-CaRING1ΔTM exhibited E3 ubiquitin ligase activity, indicating that the RING-H2 domain is essential for enzyme activity. [See online article for color version of this figure.]