Isoprenoid biosynthesis is one of the essential metabolic pathways in plants and other organisms. Despite the importance of isoprenoids for plant functions, not much is known about the regulation of isoprenoid synthesis. Quantitative technologies and systems approaches are now increasingly used to investigate the regulation of metabolic pathways and networks. A prerequisite for systems approaches is the knowledge of network elements and topologies. Information that can be extracted from the public metabolic pathway databases such as AraCyc (http://www.arabidopsis.org/biocyc) and Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg) often is not sufficiently comprehensive and current. Therefore, we built a database of manually curated isoprenoid pathway models and genes, the Arabidopsis thaliana Isoprenoid Pathway Database (AtIPD; http://www.atipd.ethz.ch). The database was compiled using information on pathways and pathway genes from BioPathAt (Lange and Ghassemian, 2003, 2005), KEGG, AraCyc, SUBA (http://suba.plantenergy.uwa.edu.au), and from the literature. AtIPD can be searched or browsed to extract data and external links related to isoprenoid pathway models, enzyme activities, or subcellular enzyme localizations. To display quantitative gene-related data on curated pathway models, we created image annotation and mapping files for integrated use with the MapMan tool (http://mapman.gabipd.org/web/guest/mapman). Additionally, we built SBML XML files of the isoprenoid pathway images using the Cell Designer tool (http://www.celldesigner.org). Users can download all image and annotation files for customization, such as adding pathway structural and regulatory network elements or modifying pathway images to visualize other quantitative protein or metabolite data. AtIPD therefore represents a valuable resource for isoprenoid network analysis.
INTRODUCTION AND STATE OF THE ART
Isoprenoid compounds are required in primary and secondary metabolic processes. As primary metabolites, they function in photosynthesis (carotenoids, chlorophylls, and plastoquinone), respiration (ubiquinone), membrane fluidity (sterols), and regulation of growth and development (cytokinins, brassinosteroids, GAs, abscisic acid, and strigolactones). As secondary metabolites, they have roles in plant protection against pathogens, in attracting pollinators and seed-dispersing animals, and in allelopathic interactions (Rodríguez-Concepción, 2006). Despite the importance of isoprenoids for plant functions, relatively little is known about the regulation of isoprenoid synthesis.
During the last few years, systems approaches to build biochemical pathway networks have become possible based on available large-scale genome, transcriptome, and metabolome data. Such studies reveal the complexity of pathway regulation and flux dynamics. Prerequisite for systems analysis of metabolic pathway regulation is the knowledge of network elements and topologies. At present, genome sequences provide the most comprehensive information to define the biological system and its metabolic pathway components. For Arabidopsis (Arabidopsis thaliana), there are currently three databases available from which information on isoprenoid pathway genes can be obtained: BioPathAt, AraCyc, and KEGG. These three databases differ in their level of comprehensiveness and annotation quality.
BioPathAt has the best-annotated isoprenoid pathways and genes (Lange and Ghassemian, 2003, 2005) because the database was curated by experts in the field, but unfortunately it has not been updated since 2005. AraCyc (http://www.arabidopsis.org/biocyc) and KEGG (http://www.genome.jp/kegg) are public online databases that were created automatically by homology searches between Arabidopsis enzymes and those from existing reference pathways. AraCyc used MetaCyc, a database of curated metabolic pathways obtained from the literature, as a source of reference pathways (Mueller et al., 2003). Reference metabolic pathways in KEGG were primarily organized from the compilations of the Japanese Biochemical Society (Nishizuka, 1980, 1997) and the wall chart of Boehringer Mannheim (Gerhard, 1992).
AraCyc was originally built by matching gene annotations (names of gene products) with enzyme names in reference pathways. The genes were then assigned to the reference pathway (Mueller et al., 2003). In KEGG, the Arabidopsis enzyme genes were annotated based on sequence similarity and positional correlation of genes. Enzyme Commission numbers and KEGG orthology codes were assigned to annotated genes. The pathway was then constructed computationally by correlating genes/sequences in the genome with gene products (enzymes) in the reference pathways according to the matching KEGG orthology code (Masoudi-Nejad et al., 2007). In both AraCyc and KEGG databases, pathway gaps (reactions with no Arabidopsis enzyme identified) were filled computationally using different algorithms (Ogata et al., 1999; Green and Karp, 2004).
Initially, the reference pathway databases used to build AraCyc and KEGG contained mainly metabolic pathways from bacteria, yeast, and mammals. As a consequence, both Arabidopsis databases primarily contained annotated genes and maps of primary metabolism. Later, AraCyc and KEGG underwent several rounds of computational and human curations, resulting in a substantial increase in the number of pathways and genes present in the metabolic pathway database. Although comprehensiveness and annotation quality of both databases improved dramatically, the list of well-annotated pathways and genes is not complete and requires additional curation prior to use. For example, certain pathways are still not annotated, such as the diterpenoid biosynthetic pathway, the apocarotenoid biosynthetic pathway, and the plastochromanol biosynthetic pathway. In case of annotated pathways, many steps lack enzymes for a given reaction. Not all reactions represent current pathway models and not all enzymes, such as prenyltransferases that share a high degree of sequence similarity, are correctly annotated to corresponding reactions and pathways. Many annotated pathway genes are pseudogenes or code for nonfunctional proteins. Often there is a redundancy in pathway models (especially in the AraCyc database) as different models are retained in the database. For example, three alternative pathways for brassinosteroid biosynthesis exist in the AraCyc database (Ephritikhine et al., 1999; Noguchi et al., 1999; Noguchi et al., 2000) instead of one preferred model (Ohnishi et al., 2006). This redundant information is confusing rather than informative. Another source of redundancy is inherent to the philosophy that is behind the construction of both databases. The majority of the pathways found in AraCyc and KEGG have some measure of experimental support, but additional computationally predicted pathways or pathway routes are also included to maximize the hypothesis-generating power of the database. These pathways or pathway routes are known to exist in other species or in nonplant organisms but have not been reported in Arabidopsis. Although this type of redundancy is advantageous in discovery-oriented research, it does not help system-oriented research in which a system and its elements should reflect reality as closely as possible.
As a first step toward systems analysis of isoprenoid pathways and their integration into the cellular biochemical network, we manually curated information from public databases and from the literature to construct AtIPD, a high-quality database of Arabidopsis isoprenoid pathways and genes. AtIPD is available online at http://www.atipd.ethz.ch and allows users to search and browse information related to isoprenoid pathways and genes and to download the genes for relation-based network analysis and pathway images. Annotation files and the mapping file can be used with the MapMan tool (http://mapman.gabipd.org/web/guest/mapman; Thimm et al., 2004) to visualize gene expression data. In addition, SBML XML files of pathway images can be downloaded and used by other programs or computer tools.
METHODOLOGY
To obtain a list of isoprenoid pathway genes (Supplemental Table S1), we compiled information on pathways and pathway genes from BioPathAt (Lange and Ghassemian, 2003, 2005), from the KEGG (http://www.genome.jp/kegg) and AraCyc (http://www.arabidopsis.org/biocyc) databases, and from the literature. We used BioPathAt as a starting point and replaced old models when more updated pathway models were found in the literature or in other databases. Novel genes were added when their function or their homology to functional proteins had been demonstrated either in the literature or in case of genes annotated by homology, at least in The Arabidopsis Information Resource (http://www.arabidopsis.org). In the last case, sequence alignments were inspected manually for the quality of hits. Activity of all gene products was verified in the literature, and the most relevant references are listed in Supplemental Table S1. As the most relevant evidence for gene function, we considered in vitro activity assays, followed by genetic complementation, metabolic profiles of mutant or overexpression lines supplemented with labeled precursors, and metabolic profiles of mutant or overexpression lines. Accordingly, genes annotated by The Arabidopsis Information Resource as expressed pseudogenes or genes encoding proteins whose function could not be confirmed experimentally were removed from the database.
Plant cells synthesize isoprenoids in the cytoplasm and different organelles. Information on the subcellular localization of individual enzymes is therefore essential to understand pathway topologies. Subcellular localization was assigned based on published in vivo and in vitro localization data (GFP fusion proteins and organelle import) and on SUBA predictions (Arabidopsis Subcellular Database; http://suba.plantenergy.uwa.edu.au; Heazlewood et al., 2007). In case of contradictory results from various resources, localization was assigned in the following order of priority: in vivo and in vitro assays, mass spectrometry data, and in silico predicted localization based on a majority vote from different prediction tools in SUBA. When enzyme localization did not comply with the predicted pathway localization and could not be confirmed by either GFP fusion protein or organelle import assays, the enzyme was assigned to the compartment in which the other pathway enzymes were localized (Supplemental Table S1, subcellular localization).
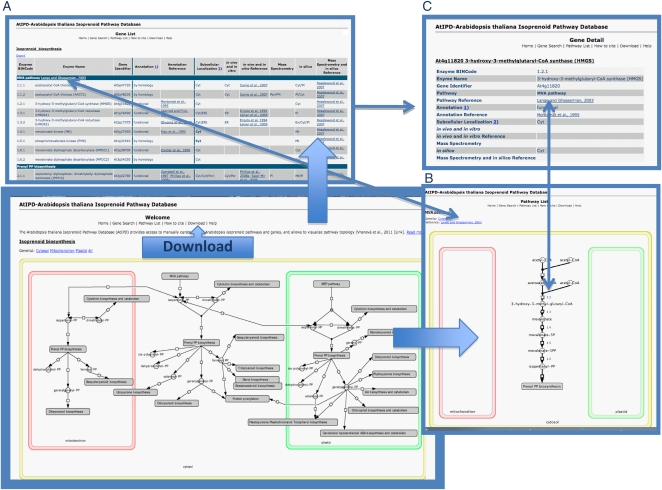
To store and visualize information on isoprenoid metabolic pathways and genes, we created a comprehensive online database that can be accessed via http://www.atipd.ethz.ch (Fig. 1). All information on genes and pathways can be searched and browsed online but can be also exported in TXT format. Pathway images were created with the Cell Designer 4.0.1 software, producing SBML XML files that can be downloaded from the download section of the online database (http://www.atipd.ethz.ch, Download, Pathway name_CD.xml). For integrated use with the MapMan tool (http://mapman.gabipd.org/web/guest/mapman), pathway images were saved as JPG files and annotated with the ImageAnnotator tool in MapMan (version 3.1.1). Genes were assigned to individual reactions using BINCodes from self-made Mapping File. Pathway images (Pathway_name.jpg), image annotations (Pathway_name.xml), and mapping file (Mapping File.xls) can be downloaded from the download section of the online database (http://www.atipd.ethz.ch, Download).
Figure 1.
The AtIPD database (http://www.atipd.ethz.ch) provides access to isoprenoid pathway models and genes. From the home page, one can navigate either to the Gene List (A) or to the Pathway List with pathway models (B). From each A and B, a link to Gene Details (C) is provided. In a Download section, pathway images (Pathway_name.jpg), image annotations (Pathway_name.xml), and a mapping file (Mapping File.xls) can be downloaded and used with the MapMan tool. Additionally, pathway images can be downloaded as SBML XML files (Pathway_name_CD.xml) and customized by the Cell Designer tool.
COMPARISON WITH OTHER DATABASES AND UTILITY IN ISOPRENOID RESEARCH
Our database of isoprenoid pathways and genes improves and outperforms already existing databases in two important aspects (Table I). First, our database has 279 correctly annotated isoprenoid pathway genes compared to 197 in BioPathAt, 205 in AraCyc, and 178 in KEGG. Second, we improved pathway annotation, including the annotation of three novel isoprenoid pathways. Plastochromanol biosynthesis was only recently described in Arabidopsis (Zbierzak et al., 2009). Plastochromanol is synthesized from plastoquinone and may regulate the antioxidant content in thylakoids and in the plastoquinone pool that is available for photosynthesis.
Table I. Comparison of different metabolic pathway gene databases.
Genes annotated as isoprenoid pathway genes in the BioPathAt database were extracted from BioPathAtDB_Oct2004.zip (http://www.ibc.wsu.edu/research/lange/public_folder/BioPathAt_V1_Oct2004/) and from Lange and Ghassemian (2003). AraCyc isoprenoid pathway genes were extracted from the AraCyc pathway database (AraCyc version 7.0; http://www.arabidopsis.org/biocyc/downloads.jsp). KEGG isoprenoid pathway genes were extracted from the KEGG PATHWAY maps (http://www.genome.jp/kegg/pathway.html).
| Pathway | No. of Genes |
|||
| BioPathAt | AraCyc | KEGG | AtIPD | |
| Mevalonic acid pathway | 9 | 7 | 8 | 9 |
| Prenyl diphosphate biosynthesis | 27 | 20 | 30 | 31 |
| Sterol biosynthesis | 20 | 22 | 19 | 25 |
| Brassinosteroid biosynthesis | 3 | 6 | 6 | 8 |
| Triterpenoid biosynthesis | 11 | 14 | 4 | 14 |
| Sesquiterpenoid biosynthesis | 16 | 3 | 0 | 16 |
| Diterpenoid biosynthesis | 19 | 0 | 0 | 19 |
| Protein prenylation | 7 | 2 | 0 | 7 |
| Cytokinin biosynthesis and catabolism | 0 | 17 | 17 | 25 |
| Ubiquinone biosynthesis | 4 | 2 | 4 | 4 |
| 2-C-Methyl-d-erythritol 4-phosphate pathway | 7 | 6 | 7 | 7 |
| Chlorophyll biosynthesis and catabolism | 29 | 35 | 30 | 37 |
| Carotenoid biosynthesis | 11 | 13 | 11 | 15 |
| Apocarotenoid biosynthesis | 0 | 2 | 0 | 5 |
| Abscisic acid biosynthesis and catabolism | 1 | 12 | 11 | 12 |
| Plastoquinone, plastochromanol, tocopherol biosynthesis | 5 | 7 | 7 | 7 |
| Phylloquinone biosynthesis | 7 | 9 | 5 | 9 |
| GA biosynthesis and catabolism | 16 | 22 | 15 | 23 |
| Monoterpenoid biosynthesis | 5 | 6 | 4 | 6 |
| Total | 197 | 205 | 178 | 279 |
Except for GA biosynthesis, a second pathway for diterpenoid biosynthesis was not predicted to exist in Arabidopsis (Aubourg et al., 2002; Lange and Ghassemian, 2003). Recently, however, geranyllinalool synthase was identified in Arabidopsis that synthesizes geranyllinalool from geranylgeranyl diphosphate (Herde et al., 2008). Based on a phylogenetic tree generated from the alignment of 32 Arabidopsis terpenoid synthases (TPSs) and 43 documented TPSs from 25 other plant species, diterpenoid synthase belongs to the same family as functional sesquiterpenoid synthases. It is therefore not possible to annotate diterpenoid synthase genes solely based on amino acid sequence similarity. We have therefore annotated predicted enzymes targeted to the cytosol or the mitochondria as both sesqui- and diterpenoid synthases, while those targeted to plastids were annotated as diterpenoid synthases. No farnesyl diphosphate synthase enzymes have been found in Arabidopsis plastids; therefore, it is unlikely that plastid TPSs have sesquiterpenoid synthase activity. Functional sesqui- and diterpenoid synthases were assigned to their respective pathways (Supplemental Table S1).
The apocarotenoid pathway was also newly annotated and besides ABA synthesis now also includes strigolactone biosynthesis and synthesis of other apocarotenoids. Strigolactones are a novel class of plant hormones derived from carotenoids that are synthesized via the MAX pathway and are known to regulate shoot branching (Bennett et al., 2006) and root architecture (Ruyter-Spira et al., 2011). Other apocarotenoids are less characterized and functionally mainly involved in flower scent and fruit flavor (Floss and Walter, 2009).
Our curation effort has allowed us to compile the most comprehensive isoprenoid pathway enzyme list available to date and also improved the quality of the database of isoprenoid pathways and genes. We used most recent pathway models based mainly on the literature data to define pathway topology. As a result, redundancy in pathway models and/or pathway routes that is intrinsic to AraCyc and KEGG databases has been removed. We have also improved the quality of gene annotation to avoid misannotation of genes that share a high degree of sequence similarity but catalyze different reactions, which is often the case based on computational prediction alone. By assigning enzymes to their correct compartments, we have further improved annotation of the isoprenoid biosynthesis pathways. Isoprenoid biosynthesis is compartmentalized in the cytosol, mitochondria, and plastids, and each compartment produces different end products. Since plant isoprenoids are derived from prenyl diphosphates that are synthesized by prenyl transferases in all three compartments, manual curation was necessary to clarify the association of prenyl transferases with different isoprenoid pathways depending on their subcellular localization.
There are many examples in metabolic pathway research for which well-annotated pathways and pathway genes are required. Basically, any type of relation-based gene network analysis requires well-curated input data. For example, coexpression analysis for identification of novel pathway elements, such as novel structural genes, regulatory genes, or transporters, relies on correctly annotated genes. Similarly, a search for common promoter motifs within a subset of genes is only successful if pathway genes are correctly annotated. A correctly curated metabolic pathway network is also prerequisite for any type of metabolic flux analysis.
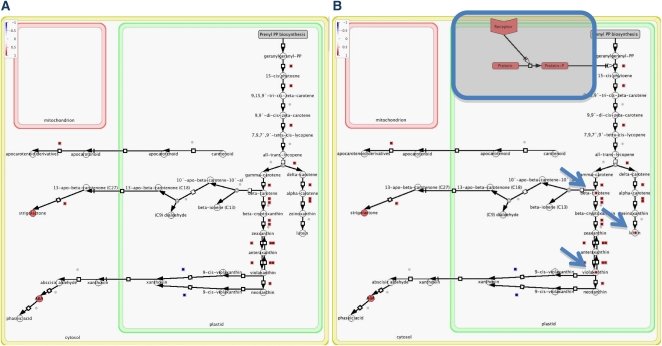
Together with a curated set of isoprenoid pathway models and genes, we also provide an online access to the data allowing easily search and browse the data, export the data, and access the information related to the isoprenoid pathway models and genes. Compared with other available pathway databases, AtIPD is the best-annotated isoprenoid gene database that is comprehensive but very user friendly and provides intuitive pathway maps and embedded information on subcellular localization and modularity. Another advantage is that both pathway images and MapMan annotation files are generated by free online tools (Cell Designer and MapMan, respectively) and can be customized by the user, thus providing an excellent tool for scientists studying the isoprenoid pathway in plants (Fig. 2). The user thus can select the pathway of interest and add pathway components, such as structural or regulatory proteins (Fig. 2A), and maps can be modified to visualize also other quantitative data, such as protein or metabolite data (Fig. 2B). Such a level of modularity is not possible with existing publicly available metabolic pathway databases.
Figure 2.
Example of data visualization on pathway images and customization of pathway images. A, Visualization of gene expression data (supplemental table II in Peschke and Kretsch, 2011; list of genes that exhibit significantly altered transcript levels at 4 h under continuous far-red light; induced +1, repressed −1) on pathway image “Carotenoid_Apocarotenoid_ABA_biosynthesis_and_catabolism.jpg” using MapMan tool and attached “Mapping_File.txt.” B, The same pathway image customized by the Cell Designer tool to add elements of a putative regulatory pathway (in frame). Additionally, images can be customized to express other gene expression data, as exemplified by visualizing metabolites (arrows). This type of customization requires annotation of other pathway elements than structural genes with the ImageAnnotator tool in MapMan and updating a mapping file (Mapping File.xls) with these new annotated elements.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. List of isoprenoid pathways and pathway genes.
Supplemental Literature Cited S1. Literature cited in Supplemental Table S1.
Acknowledgments
We thank Diana Coman, Gilles Beck, and Sean Walsh for discussions and feedback on the manuscript. Furthermore, we sincerely apologize to all colleagues whose work could not be cited because of space constraints.
References
- Aubourg S, Lecharny A, Bohlmann J. (2002) Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Genet Genomics 267: 730–745 [DOI] [PubMed] [Google Scholar]
- Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O. (2006) The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr Biol 16: 553–563 [DOI] [PubMed] [Google Scholar]
- Ephritikhine G, Pagant S, Fujioka S, Takatsuto S, Lapous D, Caboche M, Kendrick RE, Barbier-Brygoo H. (1999) The sax1 mutation defines a new locus involved in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J 18: 315–320 [DOI] [PubMed] [Google Scholar]
- Floss DS, Walter MH. (2009) Role of carotenoid cleavage dioxygenase 1 (CCD1) in apocarotenoid biogenesis revisited. Plant Signal Behav 4: 172–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard M. (1992) Biological Pathways, Ed 3 Boehringer Mannheim, Mannheim, Germany [Google Scholar]
- Green ML, Karp PD. (2004) A Bayesian method for identifying missing enzymes in predicted metabolic pathway databases. BMC Bioinformatics 5: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. (2007) SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Res (Database issue) 35: D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herde M, Gärtner K, Köllner TG, Fode B, Boland W, Gershenzon J, Gatz C, Tholl D. (2008) Identification and regulation of TPS04/GES, an Arabidopsis geranyllinalool synthase catalyzing the first step in the formation of the insect-induced volatile C16-homoterpene TMTT. Plant Cell 20: 1152–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Ghassemian M. (2003) Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol Biol 51: 925–948 [DOI] [PubMed] [Google Scholar]
- Lange BM, Ghassemian M. (2005) Comprehensive post-genomic data analysis approaches integrating biochemical pathway maps. Phytochemistry 66: 413–451 [DOI] [PubMed] [Google Scholar]
- Masoudi-Nejad A, Goto S, Endo TR, Kanehisa M. (2007) KEGG bioinformatics resource for plant genomics research. Methods Mol Biol 406: 437–458 [DOI] [PubMed] [Google Scholar]
- Mueller LA, Zhang P, Rhee SY. (2003) AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol 132: 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka T. (1980) Metabolic Maps. Biochemical Society of Japan, Tokyo [Google Scholar]
- Nishizuka T. (1997) Cell Functions and Metabolic Maps. Biochemical Society of Japan, Tokyo [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Tax FE, Yoshida S, Feldmann KA. (2000) Biosynthetic pathways of brassinolide in Arabidopsis. Plant Physiol 124: 201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Takatsuto S, Sakurai A, Yoshida S, Li J, Chory J. (1999) Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-En-3-one to (24R)-24-methyl-5α-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol 120: 833–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. (1999) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 27: 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Szatmari A-M, Watanabe B, Fujita S, Bancos S, Koncz C, Lafos M, Shibata K, Yokota T, Sakata K, et al. (2006) C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 18: 3275–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschke F, Kretsch T. (2011) Genome-wide analysis of light-dependent transcript accumulation patterns during early stages of Arabidopsis seedling deetiolation. Plant Physiol 155: 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Concepción M. (2006) Early steps in isoprenoid biosynthesis: multilevel regulation of the supply of common precursors in plant cells. Phytochem Rev 5: 1–15 [Google Scholar]
- Ruyter-Spira C, Kohlen W, Charnikhova T, van Zeijl A, van Bezouwen L, de Ruijter N, Cardoso C, Lopez-Raez JA, Matusova R, Bours R, et al. (2011) Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol 155: 721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Zbierzak AM, Kanwischer M, Wille C, Vidi PÄ, Giavalisco P, Lohmann A, Briesen I, Porfirova S, Bréhélin C, Kessler F, et al. (2009) Intersection of the tocopherol and plastoquinol metabolic pathways at the plastoglobule. Biochem J 425: 389–399 [DOI] [PubMed] [Google Scholar]