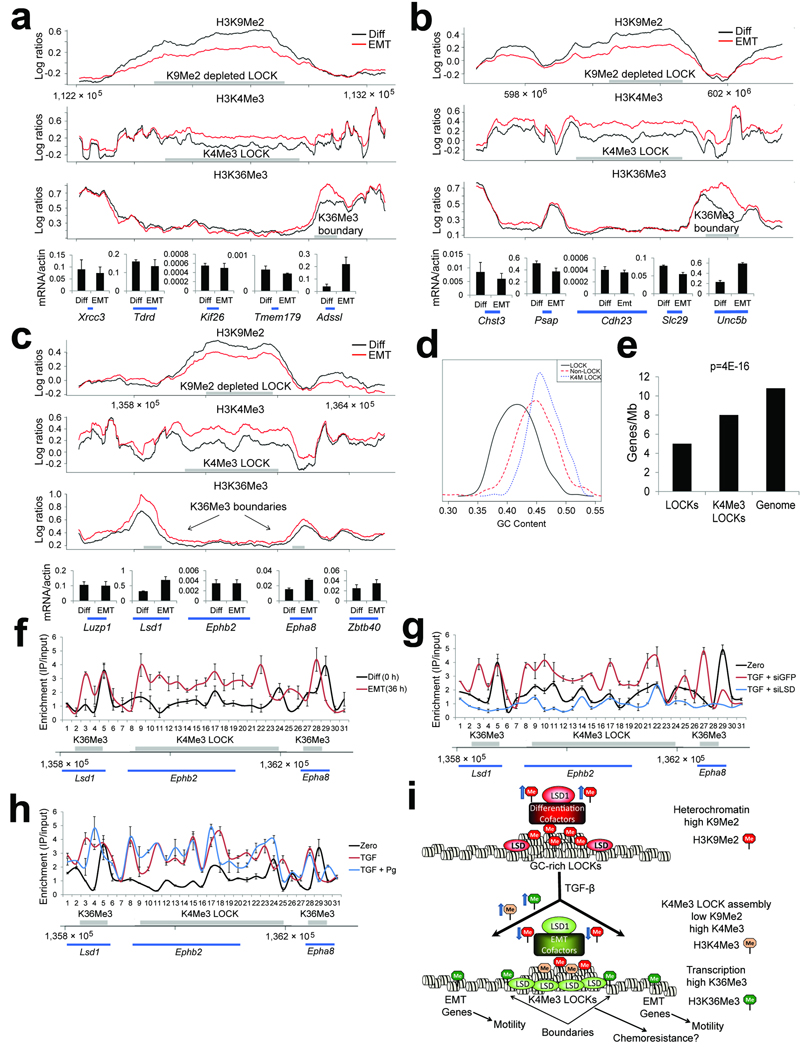
Figure 4. ChIP-chip analysis of EMT reveals alterations of chromatin in LOCK domains.
(a–c) Representative plots of ChIP-chip enrichments of histone modifications over loci from mouse chromosomes 12 (A), 10 (B) and 4 (C) during TGF-β induced EMT. Locations of genes that were assayed by RT-PCR are shown at the bottom. Regions where we detected LOCKs (top panel, H3K9Me2), K4Me3 LOCKs (middle panel, H3K4Me3), and peaks of H3K36Me3 (bottom panel, H3K36Me3) are displayed as gray bars. Chromatin was harvested from differentiated AML12 cells (time 0 hrs) and from AML12 cells undergoing EMT (TGF-β treated, 36 hrs). There is reduction in H3K9Me2 within LOCK regions, coupled with enrichment of H3K4Me3 in the same regions. Areas with enriched H3K36Me3 are present over genes at the boundaries of these LOCKs. See Fig. S8–9 for replicates of various regions, including region shown in Fig. 4C by ChIP-qPCR. Graphs underneath the ChIP-chip plots show RT-PCR results for the genes beneath them, demonstrating that genes at LOCK boundaries that acquire H3K36Me3 are upregulated during EMT.
(d) Histogram comparing GC content of LOCKs, K4Me3 LOCKs, and the whole genome. Whereas LOCKs are AT-rich overall, those that acquire H3K4Me3 during EMT are GC-rich.
(e) Gene content in all LOCKs, K4Me3 LOCKs, and the whole genome. Whereas LOCKs are gene-poor overall, the subset of LOCKs that acquire K4Me3 during EMT are gene-enriched over LOCKs that do not.
(f) ChIP assays for Lsd1 across the locus depicted in Fig. 4C. ChIP assays were performed for Lsd1 and quantitative real-time PCR was performed with 31 primers spaced across the locus. Gray bars represent where we detected a K4Me3 LOCK and H3K36Me3-enriched boundaries by ChIP-chip. Whereas Lsd1 binding is restricted to the 5’-end of the Lsd1 and Epha8 genes in differentiated cells, Lsd1 is enriched across the entire K4Me3 LOCK during EMT.
(g) ChIP assays for Lsd1 as in Fig. 4F show that chromatin isolated from AML12 cells co-treated with TGF-β and siLsd1 shows loss of Lsd1 enrichment within the K4Me3 LOCK during EMT.
(h) ChIP assays for Lsd1 as in Fig. 4F show that chromatin isolated from AML12 cells cotreated with TGF-β and pargyline shows retention of Lsd1 enrichment within the K4Me3 LOCK during EMT.
(i) Model of LOCK reprogramming during EMT. In differentiated AML12 cells, LOCKs have high levels of H3K9Me2. Lsd1 is complexed with proteins that may facilitate heterochromatin assembly at LOCKs. During TGF-β-mediated EMT, Lsd1 spreads from LOCK boundaries into the LOCKs and other proteins converge upon Lsd1, which may assist in directing demethylation of H3K9Me2 and recruitment of H3K4Me3. H3K9Me2 within all LOCKs is reduced, and specific, GC-rich LOCKs acquire H3K4Me3 adjacent to sites of transcription. These K4Me3 LOCKs might function in a surveillance DDR pathway, providing enhanced chemoresistance during EMT. H3K36Me3 is targeted to K4Me3 LOCK boundaries and numerous EMT-related genes located in non-LOCK regions across the genome, which are involved in conferring cell motility during EMT.