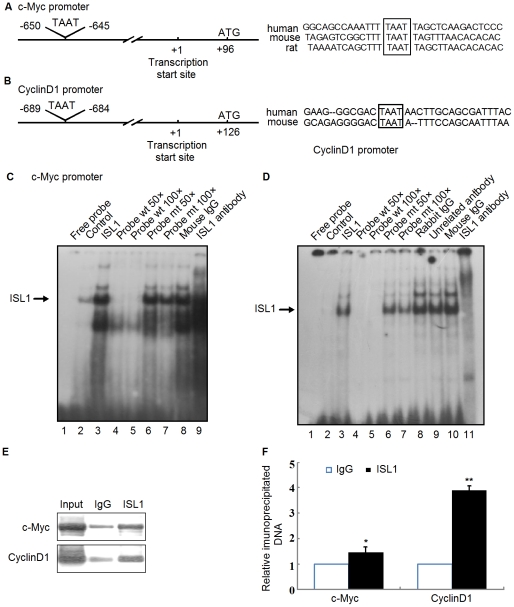
Figure 6. ISL1 band on the c-Myc or Cyclin D1 promoter directly.
Consensus binding site (TAAT, box) for ISL1 on the c-Myc (A) or CyclinD1 (B) promoter was analyzed by Matinspector software. (C and D) Nuclear extracts were subjected to EMSA for the ISL1 proteins binding ability to the 32P-labeled oligonucleotides containing the consensus sequence of the c-Myc or CyclinD1 promoter. (C) Lane 1, free probe. Lane 2, nuclear extracts from HIT-T15 cell without transfection. Lanes 3–9, nuclear extracts from HIT-T15 cells transfected with ISL1 expression construct. Lane 3 shows the direct binding of ISL1. Lanes 4 and 5 show wild-type unlabeled oligonucleotide competition. Lanes 6 and 7 show mutant unlabeled probe competition. Lane 8, the mouse normal IgG was used as a negative control. Lane 9, 1 µg anti-ISL1 antibody (H00003670-M05, Abnova) was added. (D) Lanes 1–7, same as C, Lane 8, rabbit normal IgG was used as a negative control. Lane 9, 1 µg unrelated rabbit anti-GATA4 was added. Lane 10, mouse normal IgG was used as a negative control. Lane 11, 1 µg mouse anti-ISL1 polyclonal antibody was added. (E and F): ISL1 recruited on the c-Myc or Cyclin D1 promoter was analyzed by ChIP assay. Soluble chromatin was prepared from HIT-T15 cells stably transfected with 2 µg of pcDNA3.1-ISL1 plasmid followed by immunoprecipitation with antibodies against ISL1. The DNA extractions were amplified using the primers that cover the ISL1 binding sites on the c-Myc or Cyclin D1 promoter by PCR (E) or real-time PCR (F) with normal IgG as a control. The data represent 3 independent experiments, each performed in triplicate. Each bar represents mean ± SD (**p<0.01, *p<0.05, vs. the control).