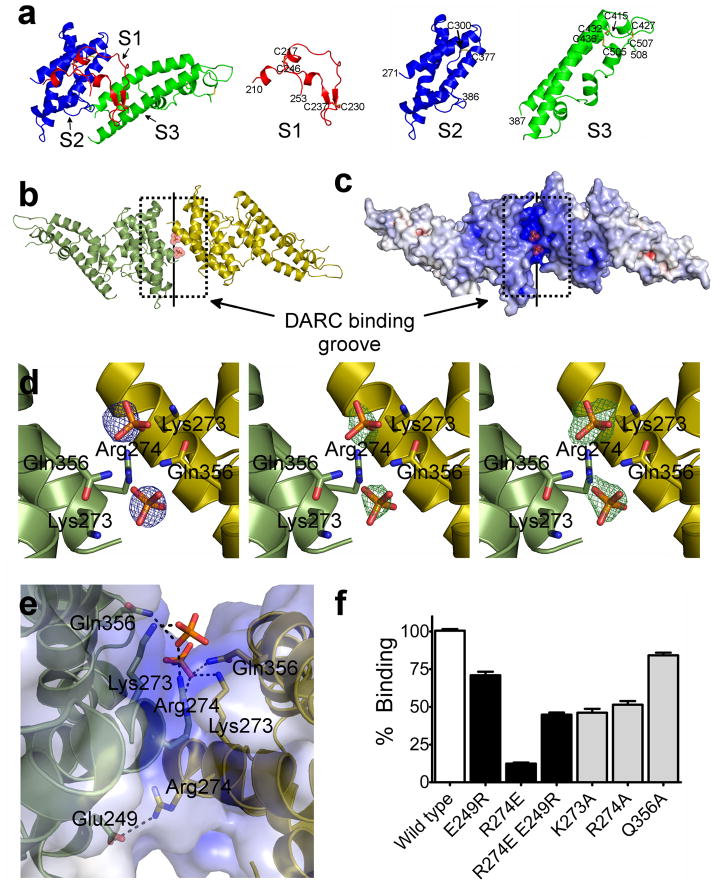
Figure 1.
RII-PvDBP is composed of three subdomains, and a sulfotyrosine pocket within a DARC binding groove is formed by the RII-PvDBP dimer. (a) RII-PvDBP separated into the three subdomains. Subdomain 1 (S1 - red) contains the β-hairpin, subdomain 2 (S2 - blue) is a four helix bundle and subdomain 3 (S3 -green) forms a second helical bundle. (b) The RII-PvDBP dimer in ribbon representation, rotated by 180° along x. Monomers are in green and yellow. The DARC binding groove is outlined by a dashed box and the dimer interface is indicated by a solid line. (c) Electrostatic mapping of the RII-PvDBP dimer, rotated by 180° along x. The DARC binding groove is positively charged. (d) Density which clearly identifies selenate or phosphate at the RII-PvDBP dimer interface is shown. Left - Selenium anomalous difference map (blue mesh) from crystals grown in the presence of selenate, contoured at 4σ. Middle - the difference map (green mesh), contoured at 2.5σ, of crystals grown in phosphate, prior to addition of phosphates. Right - the omit map (green mesh), contoured at 2.5σ, of the final refined structure with the phosphates omitted. Phosphates are drawn in stick and colored red and yellow. Side chains of residues involved in interactions are shown in stick. (e) The putative sulfotyrosine binding pocket and the Arg274-Glu249 salt bridge shown. Phosphates are drawn in stick and colored red and yellow. Side chains of residues involved in interactions are shown in stick and contacts are depicted by dashed black lines. (f) Percentage of cells expressing point mutants of RII-PvDBP that bind RBCs relative to wildtype, shown with standard error. A paired two-tailed student t-test indicated that all mutants compared to wildtype have a p-value<0.0001. White bar – wildtype. Black bars – dimer mutants and rescue. Grey bars – sulfotyrosine binding mutants.