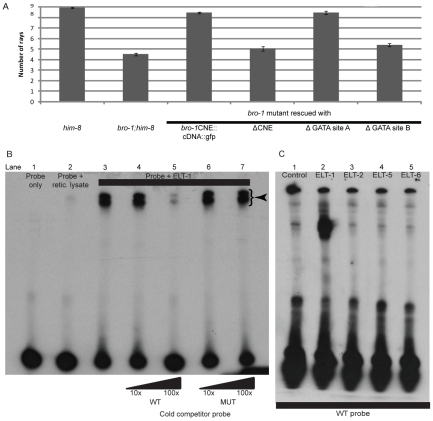
Figure 3. Correct bro-1 expression depends on ELT-1 binding to GATA site B.
(A) Bar chart showing ray number in the following strains: him-8, bro-1;him-8, bro-1;him-8; bro-1CNE::bro-1cDNA::gfp, bro-1;him-8;bro-1ΔCNE::dsRED2, bro-1;him-8;bro-1CNEΔGATA site A::bro-1cDNA::gfp, bro-1;him-8;bro-1CNEΔGATA site B::bro-1cDNA::gfp. Deletion of GATA site A has no effect on the ability of the CNE::bro-1 cDNA construct to rescue the bro-1 mutant tail phenotype, while deletion of site B abolishes the rescuing ability of the construct. Error bars show SEM. (B) EMSA showing that in vitro translated ELT-1 shifts labelled probe (lane 3, DNA-protein complexes labelled with an arrowhead), which is not shifted on its own or in the presence of reticulocyte lysate only (lanes one and two respectively). Non-labelled WT cold competitor probe at 100× concentration (lane 5) significantly reduces the intensity of the shifted bands. Addition of mutated competitor probe has no such effect at 100× concentration (lane 7), demonstrating that the ELT-1-CNE interaction is dependent on GATA site B, which is altered in the mutated probe. (C) The interaction between ELT-1 and the bro-1 CNE is specific. Incubation of ELT-1 protein with labelled probe results in a shift (lane 2) relative to the control incubation (lane 1). Incubation of other GATA transcription factors (ELT-2, ELT-5 and ELT-6) with the labelled probe resulted in no shift. ELT-3 was not tested as in vitro transcription and translation proved problematic but, like ELT-2, ELT-3 is not expressed in the seam [61] so would not bind to bro-1 in seam cells, and was not detected amongst positive clones in the yeast one-hybrid screen, despite being present in the TF library used.